6URQ
 
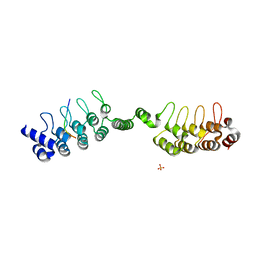 | | Complex structure of human poly-ADP-ribosyltransferase TNKS1 ARC2-ARC3 and P antigen family member 4 (PAGE4) | | Descriptor: | GLYCEROL, P antigen family member 4, Poly [ADP-ribose] polymerase tankyrase-1, ... | | Authors: | Zheng, Y, Koirala, S, Miller, D, Potts, P.R. | | Deposit date: | 2019-10-24 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tissue-Specific Regulation of the Wnt/ beta-Catenin Pathway by PAGE4 Inhibition of Tankyrase.
Cell Rep, 32, 2020
|
|
6LR8
 
 | |
7JH4
 
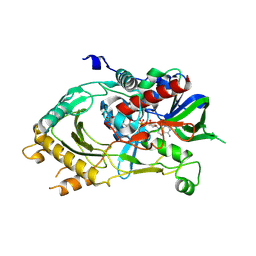
 | | Crystal structure of NAD(P)H-flavin oxidoreductase (NfoR) from S. aureus complexed with reduced FMN and NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H-dependent oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Y, O'Neill, A.G, Beaupre, B.A, Liu, D, Moran, G.R. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NfoR: Chromate Reductase or Flavin Mononucleotide Reductase?
Appl.Environ.Microbiol., 86, 2020
|
|
6JBY
 
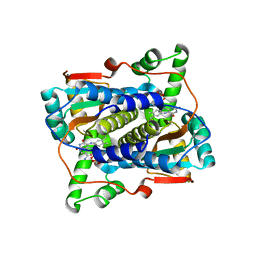
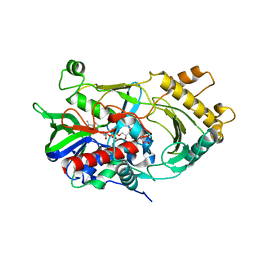 | | Crystal structure of endo-deglycosylated hydroxynitrile lyase isozyme 5 of Prunus communis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zheng, Y.C, Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2019-01-27 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Tuning of a Hydroxynitrile Lyase to Accept Rigid Pharmaco Aldehydes.
Acs Catalysis, 2020
|
|
6OJJ
 
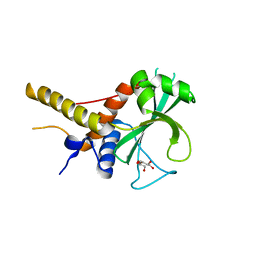 | |
4YIA
 
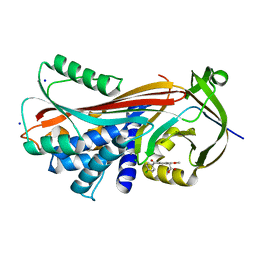 | |
6V99
 
 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase- S72D in the presence of sulfate | | Descriptor: | Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Zheng, Y, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6V96
 
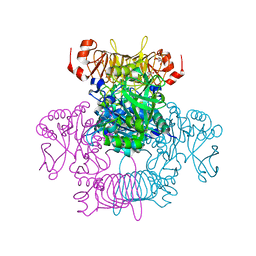 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72E | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Hussien, R, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6V9A
 
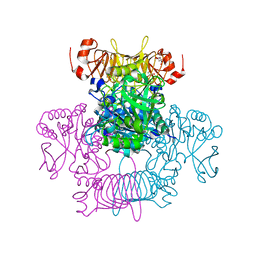 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72D | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
3WGG
 
 | | Crystal structure of RSP in complex with alpha-NAD+ | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION, alpha-Diphosphopyridine nucleotide | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding mode of the oxidized alpha-anomer of NAD(+) to RSP, a Rex-family repressor
Biochem.Biophys.Res.Commun., 456, 2015
|
|
3X2E
 
 | | A thermophilic hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Adenosylhomocysteinase | | Authors: | Zheng, Y, Ko, T.P, Huang, C.H. | | Deposit date: | 2014-12-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of S-adenosylhomocysteine hydrolase from the thermophilic bacterium Thermotoga maritima.
J.Struct.Biol., 190, 2015
|
|
7ULX
 
 | | Human DDAH1 soaked with its inhibitor N4-(4-chloropyridin-2-yl)-L-asparagine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, N-(pyridin-2-yl)-L-asparagine | | Authors: | Zheng, Y, Butrin, A, Tuley, A, Liu, D, Fast, W. | | Deposit date: | 2022-04-05 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Optimization of a switchable electrophile fragment into a potent and selective covalent inhibitor of human DDAH1
To Be Published
|
|
7ULV
 
 | | Human DDAH1 soaked with its inactivator S-((4-chloropyridin-2-yl)methyl)-L-cysteine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, S-[(pyridin-2-yl)methyl]-L-cysteine | | Authors: | Zheng, Y, Butrin, A, Tuley, A, Liu, D, Fast, W. | | Deposit date: | 2022-04-05 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Optimization of a switchable electrophile fragment into a potent and selective covalent inhibitor of human DDAH1
To Be Published
|
|
3X2F
 
 | | A Thermophilic S-Adenosylhomocysteine Hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Adenosylhomocysteinase, NITRATE ION, ... | | Authors: | Zheng, Y, Ko, T.P, Huang, C.H. | | Deposit date: | 2014-12-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structures of S-adenosylhomocysteine hydrolase from the thermophilic bacterium Thermotoga maritima.
J.Struct.Biol., 190, 2015
|
|
3S28
 
 | | The crystal structure of sucrose synthase-1 in complex with a breakdown product of the UDP-glucose | | Descriptor: | 1,5-anhydro-D-arabino-hex-1-enitol, 1,5-anhydro-D-fructose, MALONIC ACID, ... | | Authors: | Zheng, Y, Garavito, R.M. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of Sucrose Synthase-1 from Arabidopsis thaliana and Its Functional Implications.
J.Biol.Chem., 286, 2011
|
|
3S27
 
 | |
3WDJ
 
 | | Crystal structure of Pullulanase complexed with maltotetraose from Anoxybacillus sp. LM18-11 | | Descriptor: | CALCIUM ION, Type I pullulanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
3WDH
 
 | | Crystal structure of Pullulanase from Anoxybacillus sp. LM18-11 | | Descriptor: | CALCIUM ION, Type I pullulanase | | Authors: | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
3WDI
 
 | | Crystal structure of Pullulanase complexed with maltotriose from Anoxybacillus sp. LM18-11 | | Descriptor: | CALCIUM ION, Type I pullulanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
1J48
 
 | | Crystal Structure of Apo-C1027 | | Descriptor: | Apoprotein of C1027 | | Authors: | Chen, Y, Li, J, Liu, Y, Bartlam, M, Gao, Y, Jin, L, Tang, H, Shao, Y, Zhen, Y, Rao, Z. | | Deposit date: | 2001-07-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Apo-C1027 and Computer Modeling Analysis of C1027 Chromophore- Protein Complex
To be published
|
|
5JBV
 
 | | Lys27-linked triubiquitin | | Descriptor: | D-ubiquitin, NITRATE ION, Ubiquitin | | Authors: | Pan, M, Gao, S, Zheng, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-05-18 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structure of Lys27-linked triubiquitin
To Be Published
|
|
5GOD
 
 | | Lys27-linked di-ubiquitin | | Descriptor: | D-ubiquitin, MAGNESIUM ION, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
8FPX
 
 | |
5GOI
 
 | | Lys48-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOB
 
 | | Lys6-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.153 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
