5H9X
 
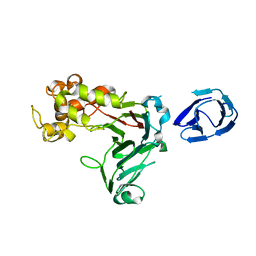 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii | | 分子名称: | beta-1,3-glucanase | | 著者 | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | 登録日 | 2015-12-29 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
5H9Y
 
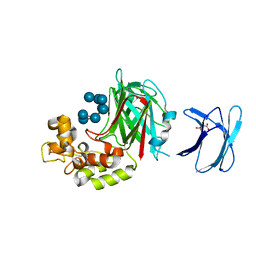 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii complexed with laminarihexaose. | | 分子名称: | L(+)-TARTARIC ACID, beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | 著者 | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | 登録日 | 2015-12-29 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.969 Å) | | 主引用文献 | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
5C3U
 
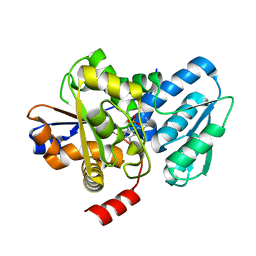 | | Crystal structure of a fungal L-serine ammonia-lyase from Rhizomucor miehei | | 分子名称: | L-serine ammonia-lyase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Zhen, Q, Qiaojuan, Y, Shaoqing, Y, Zhengqiang, J. | | 登録日 | 2015-06-17 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure and characterization of a novel l-serine ammonia-lyase from Rhizomucor miehei.
Biochem.Biophys.Res.Commun., 466, 2015
|
|
7X35
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
5XS5
 
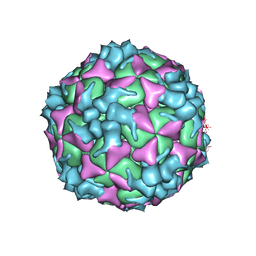 | | Structure of Coxsackievirus A6 (CVA6) virus procapsid particle | | 分子名称: | Genome polyprotein | | 著者 | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Cheng, T, Li, S.W. | | 登録日 | 2017-06-12 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS7
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle in complex with the neutralizing antibody fragment 1D5 | | 分子名称: | Genome polyprotein, Heavy chain of Fab 1D5, Light chain of Fab 1D5 | | 著者 | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | 登録日 | 2017-06-12 | | 公開日 | 2017-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS4
 
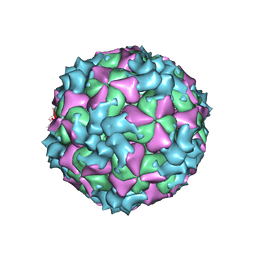 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle | | 分子名称: | Genome polyprotein | | 著者 | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | 登録日 | 2017-06-12 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
7FJ1
 
 | | Cryo-EM structure of pseudorabies virus C-capsid | | 分子名称: | Capsid vertex component 1, DNA packaging tegument protein UL25, Major capsid protein, ... | | 著者 | Zheng, Q, Li, S, Zha, Z, Sun, H. | | 登録日 | 2021-08-02 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (4.43 Å) | | 主引用文献 | Structures of pseudorabies virus capsids.
Nat Commun, 13, 2022
|
|
7FJ3
 
 | | Cryo-EM structure of PRV A-capid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Zheng, Q, Li, S, Zha, Z, Sun, H. | | 登録日 | 2021-08-02 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (4.53 Å) | | 主引用文献 | Structures of pseudorabies virus capsids.
Nat Commun, 13, 2022
|
|
6LAT
 
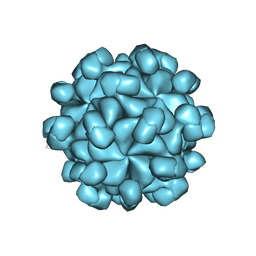 | |
6LB0
 
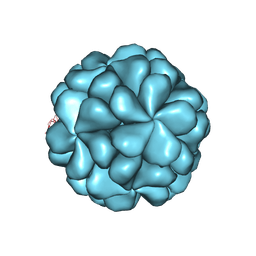 | |
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Zheng, Q, Li, S. | | 登録日 | 2019-12-05 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6AJ2
 
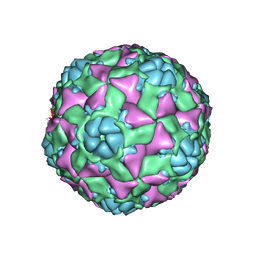 | | The structure of ICAM-5 triggered Enterovirus D68 virus A-particle | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | 著者 | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | 登録日 | 2018-08-26 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ0
 
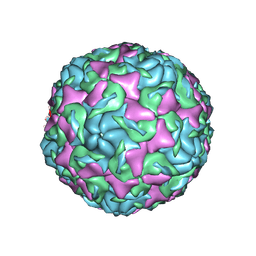 | | The structure of Enterovirus D68 mature virion | | 分子名称: | Capsid protein VP3, Capsid protein VP4, Viral protein 1, ... | | 著者 | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | 登録日 | 2018-08-25 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ9
 
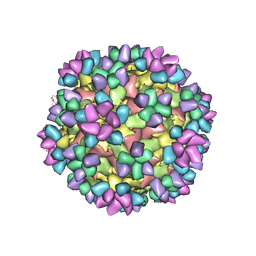 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 and 11G1 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | 登録日 | 2018-08-27 | | 公開日 | 2018-11-07 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ7
 
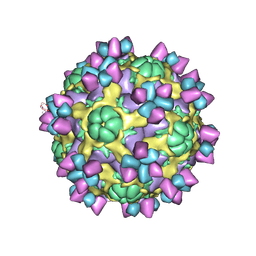 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | 登録日 | 2018-08-27 | | 公開日 | 2018-11-07 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ3
 
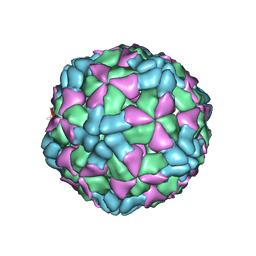 | | The structure of Enterovirus D68 procapsid | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | 著者 | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | 登録日 | 2018-08-26 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | 著者 | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | 登録日 | 2022-01-01 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WHZ
 
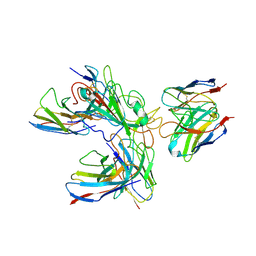 | | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | 著者 | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | 登録日 | 2022-01-01 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | 著者 | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | 登録日 | 2022-01-23 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (3.81 Å) | | 主引用文献 | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | 著者 | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | 登録日 | 2022-01-23 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7FBI
 
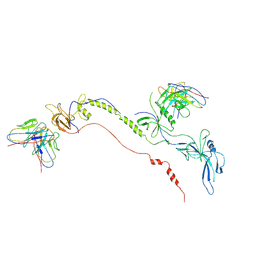 | | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5 | | 分子名称: | 3A3 heavy chain, 3A3 light chain, 3A5 heavy chain, ... | | 著者 | Zheng, Q, Li, S, Zha, Z, Hong, J, Chen, Y, Zhang, X. | | 登録日 | 2021-07-10 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5
To Be Published
|
|
7DPF
 
 | |
7X38
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 8A10 (CVB1-E:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3F
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 9A3 (CVB1-A:9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
