6OCT
 
 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3Q5K
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor | | Descriptor: | 4-[6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]-2-{[2-(methylsulfanyl)ethyl]amino}benzamide, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor
To be Published
|
|
1EV7
 
 | | CRYSTAL STRUCTURE OF DNA RESTRICTION ENDONUCLEASE NAEI | | Descriptor: | TYPE IIE RESTRICTION ENDONUCLEASE NAEI | | Authors: | Huai, Q, Colandene, J.D, Chen, Y, Luo, F, Zhao, Y. | | Deposit date: | 2000-04-19 | | Release date: | 2000-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of NaeI-an evolutionary bridge between DNA endonuclease and topoisomerase.
EMBO J., 19, 2000
|
|
3Q5J
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-{[3-(dimethylamino)propyl]amino}-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin.
To be Published
|
|
4Q79
 
 | | Structure of a HG-derivative CsgG | | Descriptor: | CsgG, MERCURY (II) ION | | Authors: | Huang, Y, Zhang, C.X, Cao, B, Zhao, Y, Kou, Y, Ni, D. | | Deposit date: | 2014-04-24 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the nonameric bacterial amyloid secretion channel
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2QER
 
 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 in the presence of dipeptide ala-pro | | Descriptor: | ALANINE, Cyclophilin-like protein, putative, ... | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Hassanali, A, Lin, L, Wasney, G, Zhao, Y, Kozieradzki, I, Vedadi, M, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 in the presence of dipeptide ala-pro.
To be Published
|
|
4OCH
 
 | | Apo structure of Smr domain of MutS2 from Deinococcus radiodurans | | Descriptor: | Endonuclease MutS2, GLYCEROL | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4001 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
2Q9Z
 
 | | Trichodiene synthase: Complex with inorganic pyrophosphate resulting from the reaction with 2-fluorofarnesyl diphosphate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Zhao, Y, Coates, R.M, Koyama, T, Cane, D.E, Christianson, D.W. | | Deposit date: | 2007-06-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Exploring biosynthetic diversity with trichodiene synthase.
Arch.Biochem.Biophys., 466, 2007
|
|
4WK9
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (0.3mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.102 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WJX
 
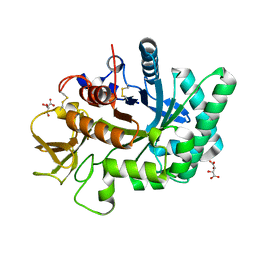 | | Crystal structure of human chitotriosidase-1 catalytic domain at 1.0 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2R0J
 
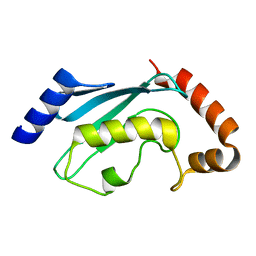 | | Crystal structure of the putative ubiquitin conjugating enzyme, PFE1350c, from Plasmodium falciparum | | Descriptor: | Ubiquitin carrier protein | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Hassanali, A, Kozieradzki, I, Zhao, Y, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the putative ubiquitin conjugating enzyme, PFE1350c, from Plasmodium falciparum.
To be Published
|
|
4WKH
 
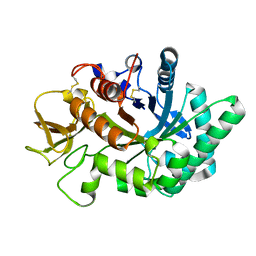 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (1mM) at 1.05 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKF
 
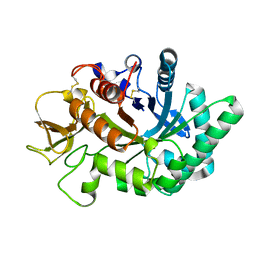 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (2.5mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKA
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain at 0.95 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2RCY
 
 | | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Ren, H, Sun, X, Khuu, C, Hassanali, A, Wasney, G, Zhao, Y, Kozieradzki, I, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound.
To be Published
|
|
2R77
 
 | | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum | | Descriptor: | Phosphatidylethanolamine-binding protein, putative | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Crombette, L, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-07 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum.
To be Published
|
|
2RMC
 
 | | Crystal structure of murine cyclophilin C complexed with immunosuppressive drug cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE C | | Authors: | Ke, H, Zhao, Y, Luo, F, Weissman, I, Friedman, J. | | Deposit date: | 1994-01-07 | | Release date: | 1995-02-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Murine Cyclophilin C Complexed with Immunosuppressive Drug Cyclosporin A
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
4N9K
 
 | | crystal structure of beta-lactamse PenP_E166S in complex with cephaloridine | | Descriptor: | 5-METHYL-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4N9L
 
 | | crystal structure of beta-lactamse PenP_E166S in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4N92
 
 | | Crystal structure of beta-lactamse PenP_E166S | | Descriptor: | Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-19 | | Release date: | 2014-09-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4OD6
 
 | | Structure of Smr domain of MutS2 from Deinococcus radiodurans, Mn2+ soaked | | Descriptor: | Endonuclease MutS2 | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
2QU8
 
 | | Crystal structure of putative nucleolar GTP-binding protein 1 PFF0625w from Plasmodium falciparum | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Putative nucleolar GTP-binding protein 1 | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Kozieradzki, I, Zhao, Y, Ravichandran, M, Shapiro, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Sukumar, D, Hassanali, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-03 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of putative nucleolar GTP-binding protein 1 PFF0625w from Plasmodium falciparum.
To be Published
|
|
3O19
 
 | | Structure-function analysis of human L-Prostaglandin D Synthase bound with fatty acid | | Descriptor: | OLEIC ACID, PALMITIC ACID, Prostaglandin-H2 D-isomerase | | Authors: | Zhou, Y, Shaw, N, Li, Y, Zhao, Y, Zhang, R, Liu, Z.-J. | | Deposit date: | 2010-07-21 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure-function analysis of human L-Prostaglandin D Synthase bound with fatty acid
To be Published
|
|
3O2Y
 
 | | Structure-function analysis of human L-Prostaglandin D Synthase bound with fatty acid | | Descriptor: | GLYCEROL, OLEIC ACID, PALMITIC ACID, ... | | Authors: | Zhou, Y, Shaw, N, Li, Y, Zhao, Y, Zhang, R, Liu, Z.-J. | | Deposit date: | 2010-07-23 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function analysis of human L-Prostaglandin D Synthase bound with fatty acid
To be Published
|
|
3TO8
 
 | | Crystal structure of the two C-terminal RRM domains of heterogeneous nuclear ribonucleoprotein L (hnRNP L) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Heterogeneous nuclear ribonucleoprotein L, ... | | Authors: | Zhang, W.J, Zeng, F.X, Liu, Y.W, Zhao, Y, Niu, L.W, Teng, M.K, Li, X. | | Deposit date: | 2011-09-04 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of the two C-terminal RRM domains of heterogeneous nuclear ribonucleoprotein L (hnRNP L)
To be Published
|
|
