3JWP
 
 | | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, TRIETHYLENE GLYCOL, Transcriptional regulatory protein sir2 homologue, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Lin, Y.H, MacKenzie, F, Senisterra, G, Allali-Hassanali, A, Vedadi, M, Ravichandran, M, Cossar, D, Kozieradzki, I, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-18 | | Release date: | 2009-10-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP
TO BE PUBLISHED
|
|
5AZC
 
 | |
5AZB
 
 | | Crystal structure of Escherichia coli Lgt in complex with phosphatidylglycerol and the inhibitor palmitic acid | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PALMITIC ACID, Prolipoprotein diacylglyceryl transferase, ... | | Authors: | Zhang, X.C, Mao, G, Zhao, Y. | | Deposit date: | 2015-09-30 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of E. coli lipoprotein diacylglyceryl transferase
Nat Commun, 7, 2016
|
|
3SI8
 
 | | Human DNA polymerase eta - DNA ternary complex with the 5'T of a CPD in the active site (TT2) | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, ... | | Authors: | Biertumpfel, C, Zhao, Y, Kondo, Y, Ramon-Maiques, S, Gregory, M, Lee, J.Y, Masutani, C, Lehmann, A.R, Hanaoka, F, Yang, W. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
3EB0
 
 | | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804 | | Descriptor: | 3-({[(3S)-3,4-dihydroxybutyl]oxy}amino)-1H,2'H-2,3'-biindol-2'-one, GLYCEROL, Putative uncharacterized protein | | Authors: | Wernimont, A.K, Fedorov, O, Lam, A, Ali, A, Zhao, Y, Lew, J, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804
TO BE PUBLISHED
|
|
3E95
 
 | | Crystal Structure of the Plasmodium Falciparum ubiquitin conjugating enzyme complex, PfUBC13-PfUev1a | | Descriptor: | UNKNOWN ATOM OR ION, Ubiquitin carrier protein, Ubiquitin-conjugating enzyme E2 | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Brokx, S, Lin, Y.H, Zhao, Y, Lew, J, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilkstrom, M, BOuntra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Qiu, W, Brand, V.B, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-21 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Plasmodium Falciparum ubiquitin conjugating enzyme complex, PfUBC13-PfUev1a
TO BE PUBLISHED
|
|
3CC9
 
 | | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Putative farnesyl pyrophosphate synthase, SODIUM ION | | Authors: | Wernimont, A.K, Dunford, J, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate.
To be Published
|
|
3CKE
 
 | | Crystal structure of aristolochene synthase in complex with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Yu, F, Miller, D.J, Faraldos, J.A, Zhao, Y, Coates, R.M, Allemann, R.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis.
J.Biol.Chem., 283, 2008
|
|
4XWW
 
 | | Crystal structure of RNase J complexed with RNA | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
3FB3
 
 | | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886 | | Descriptor: | N-acetyltransferase | | Authors: | Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886
TO BE PUBLISHED
|
|
3EZ3
 
 | | Crystal Structure of Plasmodium vivax geranylgeranylpyrophosphate synthase PVX_092040 with zoledronate and IPP bound | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Wernimont, A.K, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-22 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Crystal Structure of Plasmodium vivax geranylgeranylpyrophosphate synthase PVX_092040 with zoledronate and IPP bound
TO BE PUBLISHED
|
|
3F9R
 
 | | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370 | | Descriptor: | MAGNESIUM ION, Phosphomannomutase, SULFATE ION | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Guther, L, Shamshad, A, MacKenzie, F, Bandini, G, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370
To be Published
|
|
1EEN
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH ACETYL-D-A-D-BPA-PTYR-L-I-P-Q-Q-G | | Descriptor: | ACETIC ACID, ALA-ASP-PBF-PTR-LEU-ILE-PRO, MAGNESIUM ION, ... | | Authors: | Puius, Y.A, Zhao, Y, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition.
Biochemistry, 39, 2000
|
|
4X2A
 
 | | Crystal structure of mouse glyoxalase I complexed with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L, Li, C, Zhao, Y, Hu, X. | | Deposit date: | 2014-11-26 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In Vitro Inhibition of Glyoxalase І by Flavonoids: New Insights from Crystallographic Analysis.
Curr Top Med Chem, 16, 2016
|
|
7BEM
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-269 Vh domain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEP
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-384 and S309 Fabs | | Descriptor: | CHLORIDE ION, COVOX-384 heavy chain, COVOX-384 light chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEH
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-316 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEN
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253 and COVOX-75 Fabs | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEL
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-88 and COVOX-45 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COVOX-45 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEK
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEO
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253H55L and COVOX-75 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEI
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-150 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-150 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEJ
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
3X2R
 
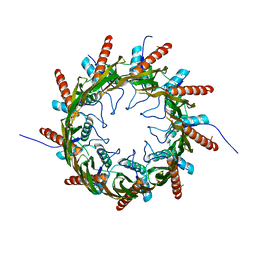 | | Structure of the nonameric bacterial amyloid secretion channel CsgG | | Descriptor: | CsgG | | Authors: | Huang, Y, Cao, B, Zhao, Y, Kou, Y, Ni, D, Zhang, X.C. | | Deposit date: | 2014-12-29 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the nonameric bacterial amyloid secretion channel
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7PJR
 
 | | Notum_ARUK3000438 | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-1,2,3-triazole, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Vecchia, L, Zhao, Y, Fish, P, Jones, E.Y. | | Deposit date: | 2021-08-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Design of a Potent, Selective, and Brain-Penetrant Inhibitor of Wnt-Deactivating Enzyme Notum by Optimization of a Crystallographic Fragment Hit.
J.Med.Chem., 65, 2022
|
|
