9C11
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36R at cryogenic temperature | | Descriptor: | CALCIUM ION, Nuclease A, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Zhang, Y, Schlessman, J.L, Siegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-05-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Domain-swapping promoted by the introduction of a charge in the hydrophobic interior of a protein
To Be Published
|
|
6GFM
 
 | | Crystal structure of the Escherichia coli nucleosidase PpnN (pppGpp-form) | | Descriptor: | Pyrimidine/purine nucleotide 5'-monophosphate nucleosidase, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Zhang, Y, Baerentsen, R.L, Gerdes, K, Brodersen, D.E. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | (p)ppGpp Regulates a Bacterial Nucleosidase by an Allosteric Two-Domain Switch.
Mol.Cell, 74, 2019
|
|
6O3Y
 
 | | Crystal structure of yeast Nrd1 CID in complex with Sen1 NIM3 | | Descriptor: | CHLORIDE ION, Helicase SEN1, Protein NRD1 | | Authors: | Zhang, Y, Tong, L. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Identification of Three Sequence Motifs in the Transcription Termination Factor Sen1 that Mediate Direct Interactions with Nrd1.
Structure, 27, 2019
|
|
6O3W
 
 | |
6GFL
 
 | |
6B40
 
 | | BbRAGL-3'TIR synaptic complex with nicked DNA refined with C2 symmetry | | Descriptor: | 31TIR intact strand, 31TIR pre-nicked strand of flanking DNA, 31TIR pre-nicked strand of signal DNA, ... | | Authors: | Zhang, Y, Cheng, T.C, Xiong, Y, Schatz, D.G. | | Deposit date: | 2017-09-25 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Transposon molecular domestication and the evolution of the RAG recombinase.
Nature, 569, 2019
|
|
6O3X
 
 | | Crystal structure of yeast Nrd1 CID in complex with Sen1 NIM2 | | Descriptor: | CHLORIDE ION, Helicase SEN1, Protein NRD1 | | Authors: | Zhang, Y, Tong, L. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Identification of Three Sequence Motifs in the Transcription Termination Factor Sen1 that Mediate Direct Interactions with Nrd1.
Structure, 27, 2019
|
|
7VWP
 
 | | Structure of the flavin-dependent monooxygenase FlsO1 from the biosynthesis of fluostatinsin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FlsO1, PHOSPHATE ION, ... | | Authors: | Zhang, Y, Yang, C, Zhang, L, Zhang, C. | | Deposit date: | 2021-11-11 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural insights of multifunctional flavin-dependent monooxygenase FlsO1-catalyzed unexpected xanthone formation
Nat Commun, 13, 2022
|
|
7Q83
 
 | |
1Z36
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z35
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
5DX4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-2-methoxybenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
1Z34
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z33
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase | | Descriptor: | purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z39
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2'-deoxyinosine | | Descriptor: | 2'-DEOXYINOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z38
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with inosine | | Descriptor: | INOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z37
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with adenosine | | Descriptor: | ADENOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
6A37
 
 | |
6P79
 
 | | Engineered single chain antibody C9+C14 ScFv | | Descriptor: | Engineered antibody heavy chain, Engineered antibody light chain | | Authors: | Zhang, Y, Li, W, Marshall, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Computer-based Engineering of Thermostabilized Antibody Fragments.
Aiche J, 66, 2020
|
|
6C00
 
 | |
6JJI
 
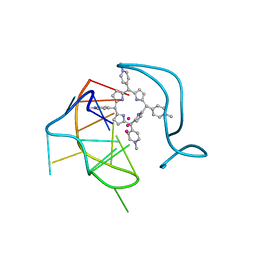 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 (1:1) | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | Authors: | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6JJH
 
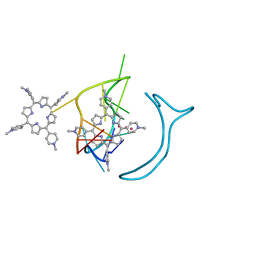 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | Authors: | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6JJF
 
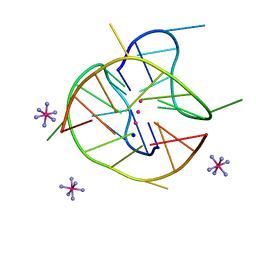 | | Crystal structure of a two-quartet DNA mixed-parallel/antiparallel G-quadruplex | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*GP*CP*TP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3'), POTASSIUM ION, ... | | Authors: | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
8CZJ
 
 | | A bacteria Zrt/Irt-like protein in the apo state | | Descriptor: | Putative membrane protein, SULFATE ION, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Zhang, Y, Hu, J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural insights into the elevator-type transport mechanism of a bacterial ZIP metal transporter.
Nat Commun, 14, 2023
|
|
8E4V
 
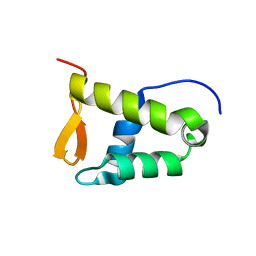 | | Solution structure of the WH domain of MORF | | Descriptor: | Isoform 3 of Histone acetyltransferase KAT6B | | Authors: | Zhang, Y, Kutateladze, T.G. | | Deposit date: | 2022-08-19 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | MORF and MOZ acetyltransferases target unmethylated CpG islands through the winged helix domain.
Nat Commun, 14, 2023
|
|
