4R2Q
 
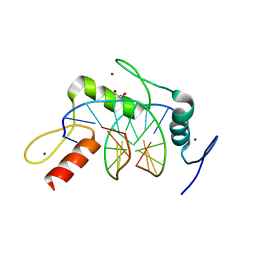 | | Wilms Tumor Protein (WT1) zinc fingers in complex with formylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5FC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5FC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
1EUE
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oganesyan, V, Zhang, X. | | Deposit date: | 2000-04-19 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of redox potential in electron transfer proteins: effects of complex formation on the active site microenvironment of cytochrome b5.
FARADAY DISC.CHEM.SOC, 116, 2001
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X5A
 
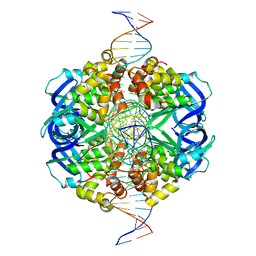 | | CryoEM structure of RuvA-Holliday junction complex | | Descriptor: | DNA (26-MER), Holliday junction ATP-dependent DNA helicase RuvA | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | Descriptor: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X7P
 
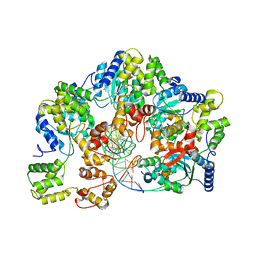 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | Descriptor: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7Q
 
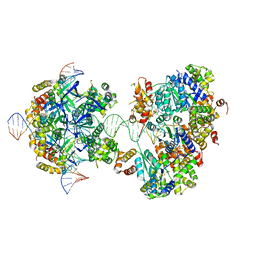 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | Descriptor: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
3L64
 
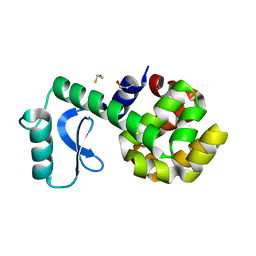 | | T4 Lysozyme S44E/WT* | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme | | Authors: | Blaber, M, Zhang, X.-J, Lindstrom, J.D, Pepiot, S.D, Baase, W.A, Matthews, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of alpha-helix propensity within the context of a folded protein. Sites 44 and 131 in bacteriophage T4 lysozyme.
J.Mol.Biol., 235, 1994
|
|
2LW6
 
 | | Solution structure of an avirulence protein AvrPiz-t from pathogen Magnaportheoryzae | | Descriptor: | AvrPiz-t protein | | Authors: | Zhang, Z.-M, Zhang, X, Zhou, Z, Hu, H, Liu, M, Zhou, B, Zhou, J. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t.
J.Biomol.Nmr, 55, 2013
|
|
8EW2
 
 | |
8EW0
 
 | |
1JRU
 
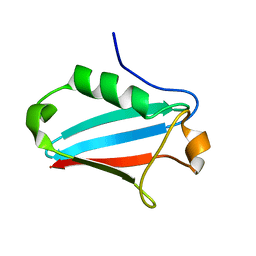 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 (ENERGY MINIMISED AVERAGE) | | Descriptor: | p47 protein | | Authors: | Yuan, X.M, Shaw, A, Zhang, X.D, Kondo, H, Lally, J, Freemont, P.S, Matthews, S.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
3CTM
 
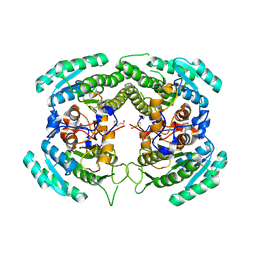 | | Crystal Structure of a Carbonyl Reductase from Candida Parapsilosis with anti-Prelog Stereo-specificity | | Descriptor: | Carbonyl Reductase | | Authors: | Zhang, R, Zhu, G, Li, X, Xu, Y, Zhang, X.C, Rao, Z. | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity.
Protein Sci., 17, 2008
|
|
7WVK
 
 | | Crystal structure of human WDR5 in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,5-bis(chloranyl)phenyl]sulfonylbenzimidazole, GLYCEROL, ... | | Authors: | Han, Q.L, Zhang, X.L, Wang, L, Ren, P.X, Cao, Y, Li, K, Bai, F. | | Deposit date: | 2022-02-10 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery, evaluation and mechanism study of WDR5-targeted small molecular inhibitors for neuroblastoma.
Acta Pharmacol.Sin., 44, 2023
|
|
2K9X
 
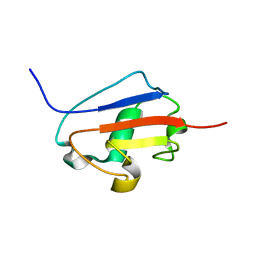 | | Solution structure of Urm1 from Trypanosoma brucei | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, W, Zhang, J, Xu, C, Wang, T, Zhang, X, Tu, X. | | Deposit date: | 2008-10-27 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 from Trypanosoma brucei
Proteins, 75, 2009
|
|
4QWQ
 
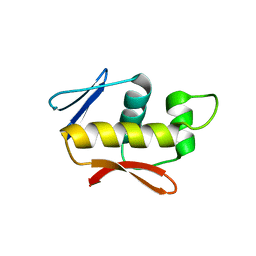 | | Crystal structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus | | Descriptor: | Response regulator SaeR | | Authors: | Fan, X, Zhu, Y, Zhang, X, Teng, M, Li, X. | | Deposit date: | 2014-07-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1WE1
 
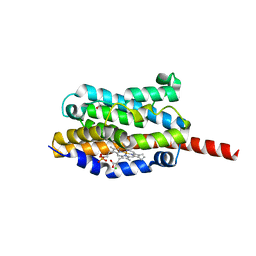 | | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC6803 in complex with heme | | Descriptor: | CHLORIDE ION, Heme oxygenase 1, ISOPROPYL ALCOHOL, ... | | Authors: | Sugishima, M, Migita, C.T, Zhang, X, Yoshida, T, Fukuyama, K. | | Deposit date: | 2004-05-21 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC 6803 in complex with heme
Eur.J.Biochem., 271, 2004
|
|
6LWA
 
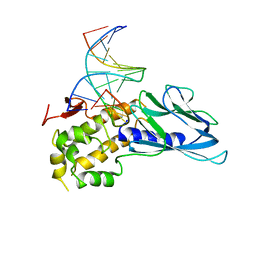 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing 5-hydroxyuracil (5-OHU) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWF
 
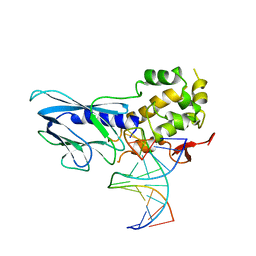 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing guanidinohydantoin (Gh) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DGH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWC
 
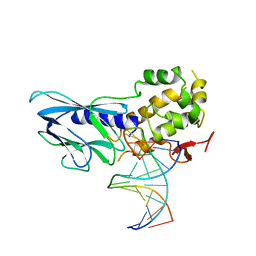 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing spiroiminodihydantoin (Sp) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DSP)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWG
 
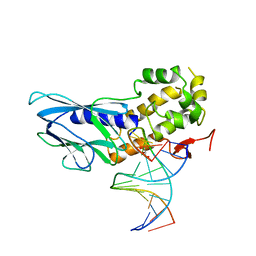 | | Crystal structure of human NEIL1(P2G, E3Q, R242) bound to duplex DNA containing guanidinohydantoin (Gh) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DGH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWB
 
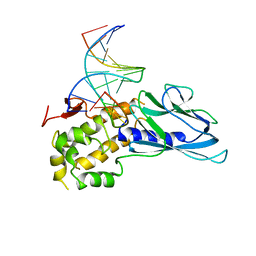 | | Crystal structure of human NEIL1(P2G, E3Q, R242) bound to duplex DNA containing 5-hydroxyuracil (5-OHU) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWN
 
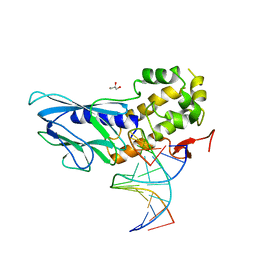 | | Crystal structure of human NEIL1(R242, G249P) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWO
 
 | | Crystal structure of human NEIL1(R242, Y244H) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWM
 
 | | Crystal structure of human NEIL1(K242) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
