7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | 分子名称: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2022-06-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y4L
 
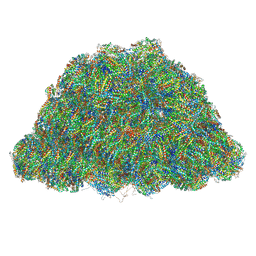 | | PBS of PBS-PSII-PSI-LHCs from Porphyridium purpureum. | | 分子名称: | Allophycocyanin alpha subunit, Allophycocyanin beta 18 subunit, Allophycocyanin beta subunit, ... | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2022-06-15 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y5E
 
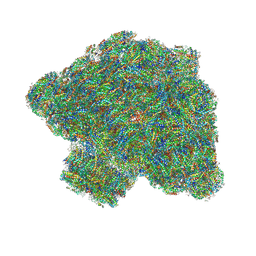 | | In situ single-PBS-PSII-PSI-LHCs megacomplex. | | 分子名称: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sui, S.F. | | 登録日 | 2022-06-17 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
8IHP
 
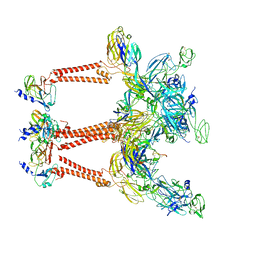 | | Structure of Semliki Forest virus VLP in complex with the receptor VLDLR-LA3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Capsid protein, ... | | 著者 | Cao, D, Ma, B, Cao, Z, Zhang, X, Xiang, Y. | | 登録日 | 2023-02-23 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of Semliki Forest virus in complex with its receptor VLDLR.
Cell, 186, 2023
|
|
7VVS
 
 | |
7W6K
 
 | | Cryo-EM structure of GmALMT12/QUAC1 anion channel | | 分子名称: | GmALMT12/QUAC1 | | 著者 | Qin, L, Tang, L.H, Xu, J.S, Zhang, X.H, Zhu, Y, Sun, F, Su, M, Zhai, Y.J, Chen, Y.H. | | 登録日 | 2021-12-01 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structure and electrophysiological characterization of ALMT from Glycine max reveal a previously uncharacterized class of anion channels.
Sci Adv, 8, 2022
|
|
7WJY
 
 | | Omicron spike trimer with 6m6 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-08 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WJZ
 
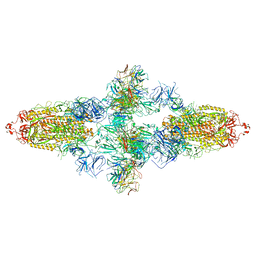 | | Omicron Spike bitrimer with 6m6 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-08 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WOG
 
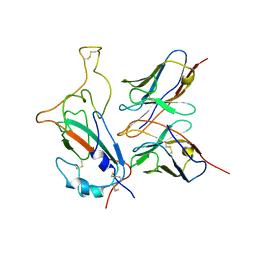 | | SARS-CoV-2 Omicron S monomer complexed with 553-49 | | 分子名称: | 553-49 VH, 553-49 VL, Spike protein S1 | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-21 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (4.06 Å) | | 主引用文献 | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WO7
 
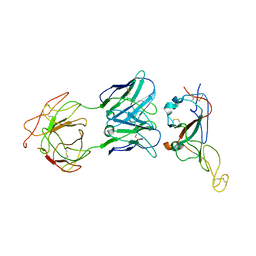 | | Locally refined region of SARS-CoV-2 Spike in complex with IgG 553-15 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, mAb15 VH, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-20 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOC
 
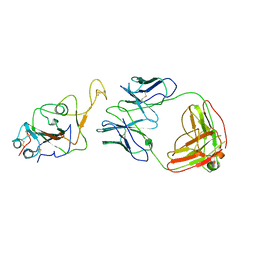 | | Locally refined region of SARS-CoV-2 Spike in complex with IgG 553-60 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, mAb60 VH, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-21 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7D0J
 
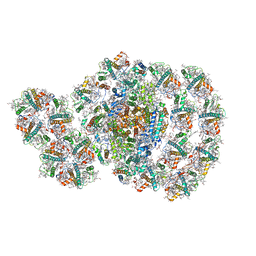 | | Photosystem I-LHCI-LHCII of Chlamydomonas reinhardtii | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Wang, W.D, Shen, L.L, Huang, Z.H, Han, G.Y, Zhang, X, Shen, J.R. | | 登録日 | 2020-09-10 | | 公開日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Structure of photosystem I-LHCI-LHCII from the green alga Chlamydomonas reinhardtii in State 2.
Nat Commun, 12, 2021
|
|
8JNT
 
 | |
7XC8
 
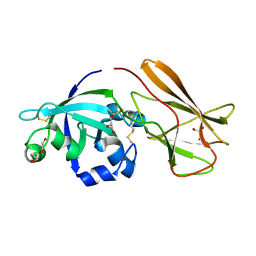 | | Crystal structure of cotton alpha-like expansin GhEXLA1 | | 分子名称: | Beta-expansin, DI(HYDROXYETHYL)ETHER, GLYCEROL | | 著者 | Zhao, F, Men, S, Xue, Y, Tu, L.L, Yin, P, Zhang, X.L. | | 登録日 | 2022-03-23 | | 公開日 | 2023-05-17 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of cotton alpha-like expansin GhEXLA1
To Be Published
|
|
7DAW
 
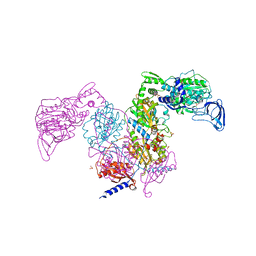 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase | | 分子名称: | Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, SULFATE ION | | 著者 | Xu, M, Zhang, X, Xu, L, Chen, S. | | 登録日 | 2020-10-18 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7DB8
 
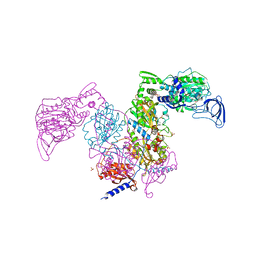 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase in complex with compound PF-3845 | | 分子名称: | N-pyridin-3-yl-4-[[3-[5-(trifluoromethyl)pyridin-2-yl]oxyphenyl]methyl]piperidine-1-carboxamide, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, ... | | 著者 | Xu, M, Zhang, X, Xu, L, Chen, S. | | 登録日 | 2020-10-19 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | 分子名称: | RNA (387-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | 分子名称: | RNA (388-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.01 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | 分子名称: | RNA (388-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | 分子名称: | RNA (388-MER) | | 著者 | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | 登録日 | 2022-05-14 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YG8
 
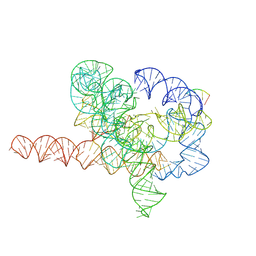 | | Cryo-EM structure of Tetrahymena ribozyme conformation 5 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (387-MER), RNA (5'-R(*CP*CP*CP*UP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YG9
 
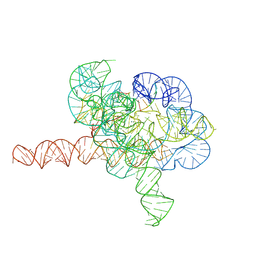 | | Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (391-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGC
 
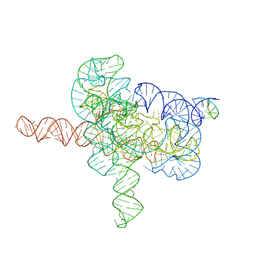 | | Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*UP*UP*AP*AP*CP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.65 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGA
 
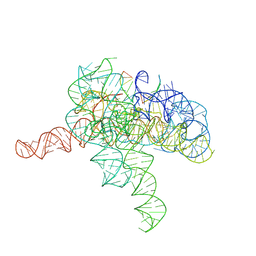 | | Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.35 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGD
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (384-MER), RNA (5'-R(*CP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
