1NN4
 
 | | Structural Genomics, RpiB/AlsB | | Descriptor: | Ribose 5-phosphate isomerase B | | Authors: | Zhang, R.G, Andersson, C.E, Mowbray, S.L, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S.L, Arrowsmith, C, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction.
J.Mol.Biol., 332, 2003
|
|
1I6N
 
 | | 1.8 A Crystal structure of IOLI protein with a binding zinc atom | | Descriptor: | IOLI PROTEIN, ZINC ION | | Authors: | Zhang, R.G, Dementiva, I, Collart, F, Quaite-Randall, E, Joachimiak, A, Alkire, R, Maltsev, N, Korolev, O, Dieckman, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-03-02 | | Release date: | 2002-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus subtilis ioli shows endonuclase IV fold with altered Zn binding.
Proteins, 48, 2002
|
|
1K4N
 
 | | Structural Genomics, Protein EC4020 | | Descriptor: | Protein EC4020 | | Authors: | Zhang, R.G, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-08 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved protein YecM from Escherichia coli shows structural homology to metal-binding isomerases and oxygenases.
Proteins, 51, 2003
|
|
2I79
 
 | | The crystal structure of the acetyltransferase of GNAT family from Streptococcus pneumoniae | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, GNAT family | | Authors: | Zhang, R.G, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the acetyltransferase of GNAT family from Streptococcus pneumoniae
To be Published
|
|
4Y21
 
 | | Crystal Structure of Munc13-1 MUN domain | | Descriptor: | Protein unc-13 homolog A | | Authors: | Yang, X.Y, Wang, S, Sheng, Y, Zhang, M, Zou, W.J, Wu, L.J, Kang, L.J, Rizo, J, Zhang, R.G, Xu, T, Ma, C. | | Deposit date: | 2015-02-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1MJN
 
 | | Crystal Structure of the intermediate affinity aL I domain mutant | | Descriptor: | Integrin alpha-L, MAGNESIUM ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.H, Yang, Y.T, Dong, Y.C, Jun, C.D, McCormack, A, Zhang, R.G, Wang, J.H, Springer, T.A. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the alphaL I Domain and its Complex with ICAM-1 reveal a Shape-shifting Pathway for Integrin Regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MC2
 
 | | monomeric LYS-49 phospholipase A2 homologue purified from AG | | Descriptor: | Acutohaemonlysin, ISOPROPYL ALCOHOL | | Authors: | Liu, Q, Huang, Q.Q, Zhang, R.G, Weeks, C.M, Jelsch, C, Teng, M.K, Niu, L.W. | | Deposit date: | 2002-08-05 | | Release date: | 2002-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The crystal structure of a novel, inactive, lysine 49 PLA2 from Agkistrodon acutus venom: an ultrahigh resolution, AB initio structure determination
J.Biol.Chem., 278, 2003
|
|
1MKM
 
 | | CRYSTAL STRUCTURE OF THE THERMOTOGA MARITIMA ICLR | | Descriptor: | FORMIC ACID, IclR transcriptional regulator, ZINC ION | | Authors: | Kim, Y, Zhang, R.G, Joachimiak, A, Skarina, T, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Thermotoga maritima 0065, a member of the IclR transcriptional factor family.
J.Biol.Chem., 277, 2002
|
|
1G8E
 
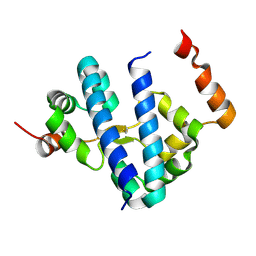 | | CRYSTAL STRUCTURE OF FLHD FROM ESCHERICHIA COLI | | Descriptor: | FLAGELLAR TRANSCRIPTIONAL ACTIVATOR FLHD | | Authors: | Campos, A, Zhang, R.G, Alkire, R.W, Matsumura, P, Westbrook, E.M. | | Deposit date: | 2000-11-17 | | Release date: | 2001-03-07 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the global regulator FlhD from Escherichia coli at 1.8 A resolution.
Mol.Microbiol., 39, 2001
|
|
5C8S
 
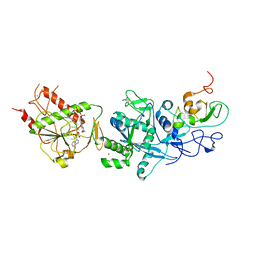 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligands SAH and GpppA | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, Guanine-N7 methyltransferase, MAGNESIUM ION, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2015-08-12 | | Method: | X-RAY DIFFRACTION (3.326 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5C8T
 
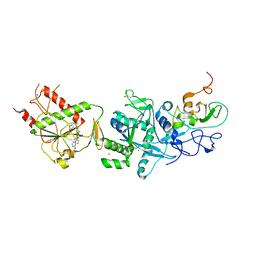 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligand SAM | | Descriptor: | Guanine-N7 methyltransferase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5C8U
 
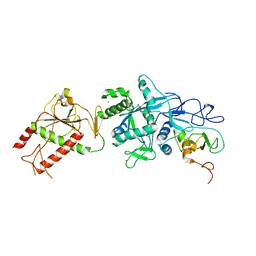 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex | | Descriptor: | Guanine-N7 methyltransferase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1XG8
 
 | | Crystal Structure of Protein of Unknown Function SA0789 from Staphylococcus aureus | | Descriptor: | hypothetical protein SA0798 | | Authors: | Rotella, F.J, Zhang, R.G, Kim, Y, Quartey, P, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A crystal structure of hypothetical protein SA0798 from Staphylococcus aureus
To be Published
|
|
1YXY
 
 | | Crystal Structure of putative N-acetylmannosamine-6-P epimerase from Streptococcus pyogenes (APC29713) Structural genomics, MCSG | | Descriptor: | Putative N-acetylmannosamine-6-phosphate 2-epimerase | | Authors: | Rotella, F.J, Zhang, R.G, Lezondra, L.E.O, Collart, F.R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of putative N-acetylmannosamine-6-P epimerase from Streptococcus pyogenes
To be Published
|
|
1YOC
 
 | | Crystal Structure of genomics APC5556 | | Descriptor: | GLYCEROL, hypothetical protein PA1835 | | Authors: | Dong, A, Evdokimova, E, Kudritskam, M, Zhang, R.G, Yakunin, A, Pai, E, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-27 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of genomics AFPA1835 by Sulfur SAD methods
To be Published
|
|
1YZE
 
 | | Crystal structure of the N-terminal domain of USP7/HAUSP. | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Saridakis, V, Sheng, Y, Sarkari, F, Holowaty, M.N, Shire, K, Nguyen, T, Zhang, R.G, Liao, J, Lee, W, Edwards, A.M, Arrowsmith, C.H, Frappier, L. | | Deposit date: | 2005-02-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.
Mol.Cell, 18, 2005
|
|
1LJ9
 
 | | The crystal structure of the transcriptional regulator SlyA | | Descriptor: | transcriptional regulator SlyA | | Authors: | Wu, R.Y, Zhang, R.G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-04-19 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor
J.Biol.Chem., 278, 2003
|
|
3OVR
 
 | | Crystal Structure of hRPE and D-Xylulose 5-Phosphate Complex | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, 5-O-phosphono-D-xylulose, FE (II) ION, ... | | Authors: | Liang, W.G, Ouyang, S.Y, Shaw, N, Joachimiak, A, Zhang, R.G, Liu, Z.J. | | Deposit date: | 2010-09-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Conversion of D-ribulose 5-phosphate to D-xylulose 5-phosphate: new insights from structural and biochemical studies on human RPE
Faseb J., 25, 2011
|
|
3OVQ
 
 | | Crystal Structure of hRPE and D-Ribulose-5-Phospate Complex | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, FE (II) ION, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Liang, W.G, Ouyang, S.Y, Shaw, N, Joachimiak, A, Zhang, R.G, Liu, Z.J. | | Deposit date: | 2010-09-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Conversion of D-ribulose 5-phosphate to D-xylulose 5-phosphate: new insights from structural and biochemical studies on human RPE
Faseb J., 25, 2011
|
|
3OVP
 
 | | Crystal Structure of hRPE | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, FE (II) ION, Ribulose-phosphate 3-epimerase | | Authors: | Liang, W.G, Ouyang, S.Y, Shaw, N, Joachimiak, A, Zhang, R.G, Liu, Z.J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Conversion of D-ribulose 5-phosphate to D-xylulose 5-phosphate: new insights from structural and biochemical studies on human RPE
Faseb J., 25, 2011
|
|
4OI6
 
 | | Crystal structure analysis of nickel-bound form SCO4226 from Streptomyces coelicolor A3(2) | | Descriptor: | CITRIC ACID, NICKEL (II) ION, Nickel responsive protein | | Authors: | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-01-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
4OI3
 
 | | Crystal structure analysis of SCO4226 from Streptomyces coelicolor A3(2) | | Descriptor: | Nickel responsive protein | | Authors: | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-01-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
5YBE
 
 | | Structure of KANK1/KIF21A complex | | Descriptor: | 1,2-ETHANEDIOL, KIF21A, Kank1 protein | | Authors: | Weng, Z.F, Shang, Y, Yao, D.Q, Zhu, J.W, Zhang, R.G. | | Deposit date: | 2017-09-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Structural analyses of key features in the KANK1/KIF21A complex yield mechanistic insights into the cross-talk between microtubules and the cell cortex.
J. Biol. Chem., 293, 2018
|
|
5ZYS
 
 | | Structure of Nephrin/MAGI1 complex | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, Nephrin | | Authors: | Weng, Z.F, Shng, Y, Zhu, J.W, Zhang, R.G. | | Deposit date: | 2018-05-28 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis of Highly Specific Interaction between Nephrin and MAGI1 in Slit Diaphragm Assembly and Signaling.
J. Am. Soc. Nephrol., 29, 2018
|
|
6A68
 
 | | the crystal structure of rat calcium-dependent activator protein for secretion (CAPS) DAMH domain | | Descriptor: | Calcium-dependent secretion activator 1, POTASSIUM ION | | Authors: | Zhou, H, Wei, Z.Q, Yao, D.Q, Zhang, R.G, Ma, C. | | Deposit date: | 2018-06-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structural and Functional Analysis of the CAPS SNARE-Binding Domain Required for SNARE Complex Formation and Exocytosis.
Cell Rep, 26, 2019
|
|
