6LJL
 
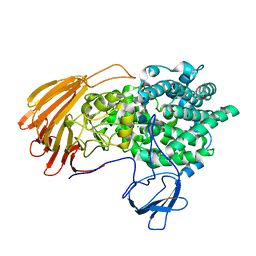 | | Crystal Structure of exoHep-Y390A/H555A complexed with a tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | Authors: | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
6M24
 
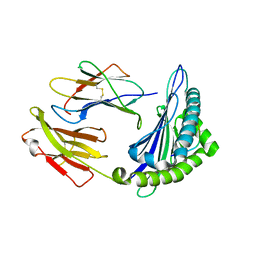 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-2 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6M2J
 
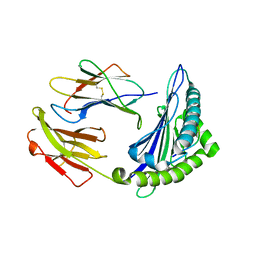 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-1 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6M2K
 
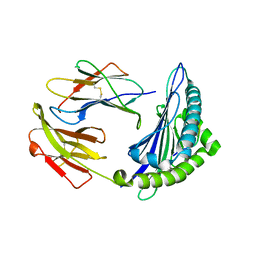 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-10 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
2H3J
 
 | | Solution NMR Structure of Protein PA4359 from Pseudomonas aeruginosa: Northeast Structural Genomics Consortium Target PaT89 | | Descriptor: | Hypothetical protein PA4359 | | Authors: | Zhang, Q, Liu, G, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Hypothetical protein PA4359: Northest Structural Genomics Target PaT89
To be Published
|
|
2NOC
 
 | | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106 | | Descriptor: | Putative periplasmic protein | | Authors: | Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106
To be Published
|
|
5DV2
 
 | |
5DV4
 
 | |
8GV9
 
 | | The cryo-EM structure of hAE2 with chloride ion | | Descriptor: | Anion exchange protein 2, CHLORIDE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVA
 
 | | The intermediate structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVE
 
 | | The asymmetry structure of hAE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GV8
 
 | | The cryo-EM structure of hAE2 with DIDS | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVC
 
 | | The cryo-EM structure of hAE2 with bicarbonate | | Descriptor: | Anion exchange protein 2, BICARBONATE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVF
 
 | | The outward-facing structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVH
 
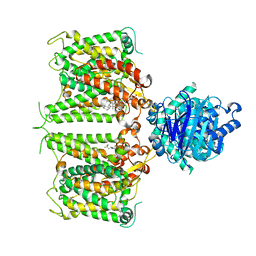 | | Human AE2 in acidic KNO3 | | Descriptor: | Anion exchange protein 2, CHOLESTEROL HEMISUCCINATE | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8V0P
 
 | |
8UK1
 
 | |
8V0T
 
 | |
8V0O
 
 | |
8V0M
 
 | |
8V0L
 
 | |
8UIR
 
 | |
8V0S
 
 | |
8V0V
 
 | |
8V0X
 
 | |
