7WWU
 
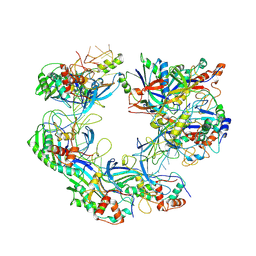 | | ICP1 Csy complex | | Descriptor: | Csy1, Csy2, Csy3, ... | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YKG
 
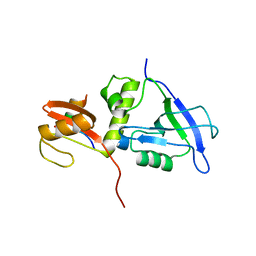 | | Crystal structure of MAGI2 PDZ0-GK/pSGEF complex | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, SGEF | | Authors: | Zhang, M, Lin, L, Zhu, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.
Sci Adv, 9, 2023
|
|
7YKI
 
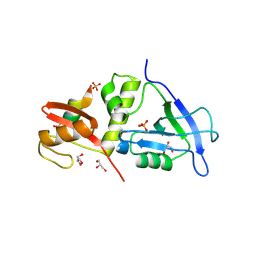 | | Crystal structure of MAGI2 PDZ0-GK domain in complex with phospho-SAPAP1 GBR3 peptide | | Descriptor: | GLYCEROL, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, ... | | Authors: | Zhang, M, Lin, L, Zhu, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.
Sci Adv, 9, 2023
|
|
7YKF
 
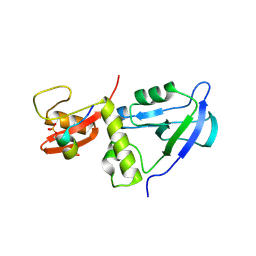 | | Crystal structure of MAGI2 PDZ0-GK/pEphexin4 complex | | Descriptor: | Ephexin4, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 | | Authors: | Zhang, M, Lin, L, Zhu, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.
Sci Adv, 9, 2023
|
|
7YKH
 
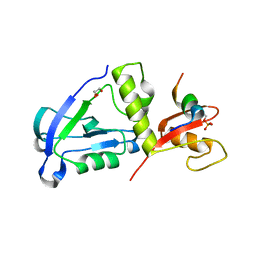 | | Crystal structure of MAGI2 PDZ0-GK domain in complex with phospho-SAPAP1 GBR2 peptide | | Descriptor: | GLYCEROL, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, ... | | Authors: | Zhang, M, Lin, L, Zhu, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphorylation-dependent recognition of diverse protein targets by the cryptic GK domain of MAGI MAGUKs.
Sci Adv, 9, 2023
|
|
6IMC
 
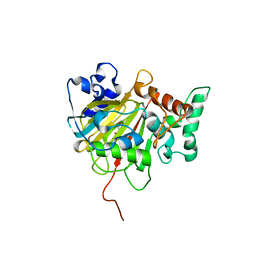 | | Crystal Structure of ALKBH1 in complex with Mn(II) and N-Oxalylglycine | | Descriptor: | MANGANESE (II) ION, N-OXALYLGLYCINE, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
6IMA
 
 | | Crystal Structure of ALKBH1 without alpha-1 (N37-C369) | | Descriptor: | CITRIC ACID, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
7CSR
 
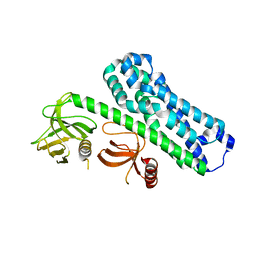 | | Structure of Ephexin4 R676L | | Descriptor: | Rho guanine nucleotide exchange factor 16 | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CSO
 
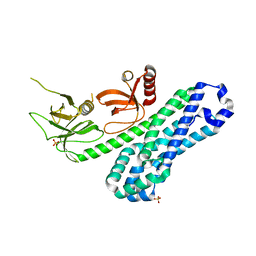 | | Structure of Ephexin4 DH-PH-SH3 | | Descriptor: | Rho guanine nucleotide exchange factor 16, SULFATE ION | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-15 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CSP
 
 | | Structure of Ephexin4 IDPSH | | Descriptor: | Rho guanine nucleotide exchange factor 16 | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-15 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1F95
 
 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND BIM PEPTIDE COMPLEX | | Descriptor: | BCL2-LIKE 11 (APOPTOSIS FACILITATOR), DYNEIN | | Authors: | Fan, J.-S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
1F96
 
 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND NNOS PEPTIDE COMPLEX | | Descriptor: | DYNEIN LIGHT CHAIN 8, PROTEIN (NNOS, NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Fan, J.S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
6DHO
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.07 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6DHG
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.5 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6DHF
 
 | | RT XFEL structure of the one-flash state of Photosystem II (1F, S2-rich) at 2.08 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6DHE
 
 | | RT XFEL structure of the dark-stable state of Photosystem II (0F, S1-rich) at 2.05 Angstrom resolution | | Descriptor: | (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
2FQD
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQE
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQG
 
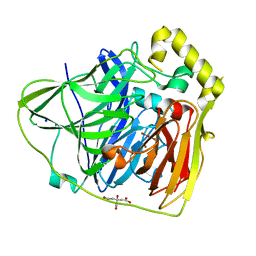 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
4Y21
 
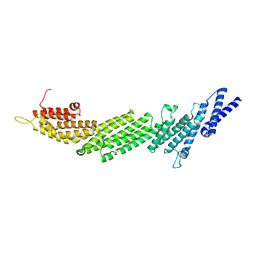 | | Crystal Structure of Munc13-1 MUN domain | | Descriptor: | Protein unc-13 homolog A | | Authors: | Yang, X.Y, Wang, S, Sheng, Y, Zhang, M, Zou, W.J, Wu, L.J, Kang, L.J, Rizo, J, Zhang, R.G, Xu, T, Ma, C. | | Deposit date: | 2015-02-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2FQF
 
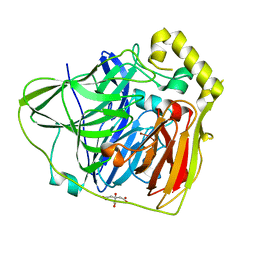 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2QT5
 
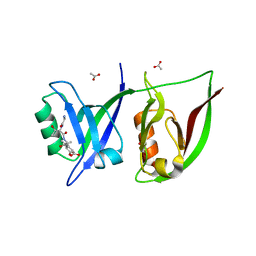 | | Crystal Structure of GRIP1 PDZ12 in Complex with the Fras1 Peptide | | Descriptor: | (ASN)(ASN)(LEU)(GLN)(ASP)(GLY)(THR)(GLU)(VAL), 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Long, J, Wei, Z, Feng, W, Zhao, Y, Zhang, M. | | Deposit date: | 2007-08-01 | | Release date: | 2008-06-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Supramodular nature of GRIP1 revealed by the structure of its PDZ12 tandem in complex with the carboxyl tail of Fras1.
J.Mol.Biol., 375, 2008
|
|
