7MSC
 
 | | Mtb 70SIC in complex with MtbEttA at Pre_R0 state | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Cui, Z, Zhang, J. | | 登録日 | 2021-05-11 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
6WB2
 
 | | +3 extended HIV-1 reverse transcriptase initiation complex core (displaced state) | | 分子名称: | HIV-1 viral RNA genome fragment, Reverse transcriptase/ribonuclease H, reverse transcriptase p51 subunit, ... | | 著者 | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Chen, D.H, Puglisi, E.V. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
8HUV
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Zeng, P, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUR
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUT
 
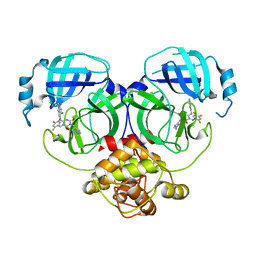 | | Crystal structure of MERS main protease in complex with S217622 | | 分子名称: | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ORF1a | | 著者 | Lin, C, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUU
 
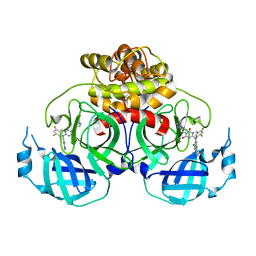 | | Crystal structure of HCoV-NL63 main protease with S217622 | | 分子名称: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Zeng, X.Y, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUS
 
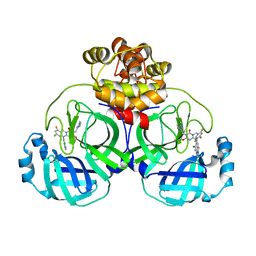 | | Crystal structure of SARS main protease in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Lin, C, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUX
 
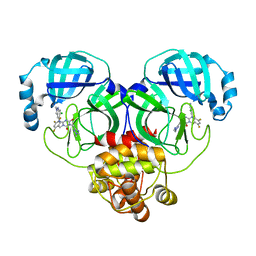 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Li, W.W, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
6WB1
 
 | | +3 extended HIV-1 reverse transcriptase initiation complex core (intermediate state) | | 分子名称: | HIV-1 viral RNA genome fragment, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, reverse transcriptase p51 subunit, ... | | 著者 | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Chen, D.H, Puglisi, E.V. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6WAZ
 
 | | +1 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | 分子名称: | HIV-1 viral RNA genome fragment, Reverse transcriptase p51 subunit, Reverse transcriptase/ribonuclease H, ... | | 著者 | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6UFM
 
 | |
2L7E
 
 | |
3IZJ
 
 | | Mm-cpn rls with ATP and AlFx | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-29 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZL
 
 | | Mm-cpn rls deltalid with ATP and AlFx | | 分子名称: | Mm-cpn rls deltalid | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-29 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
6OL3
 
 | | Crystal structure of an adenovirus virus-associated RNA | | 分子名称: | Adenovirus Virus-Associated (VA) RNA I apical and central domains, POTASSIUM ION | | 著者 | Hood, I.V, Gordon, J.M, Bou-Nader, C, Henderson, F.V, Bahmanjah, S, Zhang, J. | | 登録日 | 2019-04-15 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Crystal structure of an adenovirus virus-associated RNA.
Nat Commun, 10, 2019
|
|
3IZN
 
 | | Mm-cpn deltalid with ATP | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-30 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZM
 
 | | Mm-cpn wildtype with ATP | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-30 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZK
 
 | | Mm-cpn rls deltalid with ATP | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-29 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
8HDK
 
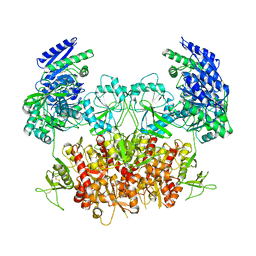 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (minor class in symmetry) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | 著者 | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | 登録日 | 2022-11-04 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3IZI
 
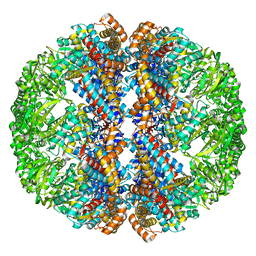 | | Mm-cpn rls with ATP | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-29 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZH
 
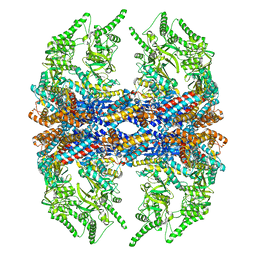 | | Mm-cpn D386A with ATP | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-29 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3GGQ
 
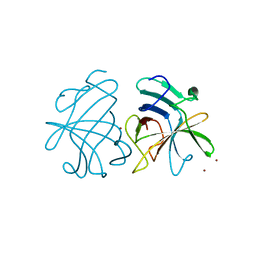 | | Dimerization of Hepatitis E Virus Capsid Protein E2s Domain is Essential for Virus-Host Interaction | | 分子名称: | BROMIDE ION, Capsid protein | | 著者 | Li, S.W, Tang, X.H, Seetharaman, J, Yang, C.Y, Gu, Y, Zhang, J, Du, H.L, Shih, J.W.K, Hew, C.L, Sivaraman, J, Xia, N.S. | | 登録日 | 2009-03-02 | | 公開日 | 2009-08-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction
Plos Pathog., 5, 2009
|
|
6LWF
 
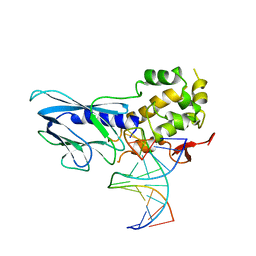 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing guanidinohydantoin (Gh) | | 分子名称: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DGH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | 著者 | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2020-02-07 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWC
 
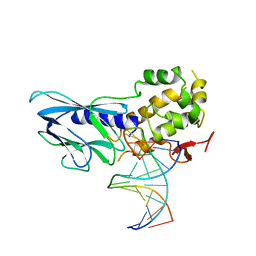 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing spiroiminodihydantoin (Sp) | | 分子名称: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DSP)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | 著者 | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2020-02-07 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWA
 
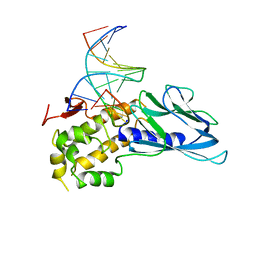 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing 5-hydroxyuracil (5-OHU) | | 分子名称: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | 著者 | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2020-02-07 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
