1U6D
 
 | |
4U92
 
 | | Crystal structure of a DNA/Ba2+ G-quadruplex containing a water-mediated C-tetrad | | Descriptor: | BARIUM ION, DNA (5'-D(*CP*CP*AP*KP*GP*CP*GP*TP*GP*G)-3'), MAGNESIUM ION | | Authors: | Paukstelis, P.J, Zhang, D, Huang, T, Lukeman, P. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a DNA/Ba2+ G-quadruplex containing a water-mediated C-tetrad.
Nucleic Acids Res., 42, 2014
|
|
8D0A
 
 | | Crystal structure of human USP30 in complex with a covalent inhibitor 829 and a Fab | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 30, ZINC ION, mouse anti-huUSP30 Fab heavy chain, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | TBD
To Be Published
|
|
4RIP
 
 | |
4M3Q
 
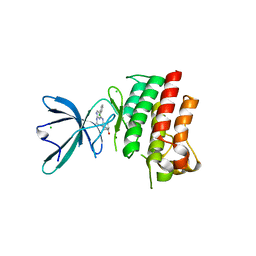 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1917 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Zhang, W, Zhang, D, Stashko, M.A, DeRyckere, D, Hunter, D, Kireev, D.B, Miley, M, Cummings, C, Lee, M, Norris-Drouin, J, Stewart, W.M, Sather, S, Zhou, Y, Kirkpatrick, G, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Pseudo-Cyclization through Intramolecular Hydrogen Bond Enables Discovery of Pyridine Substituted Pyrimidines as New Mer Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
7X1Z
 
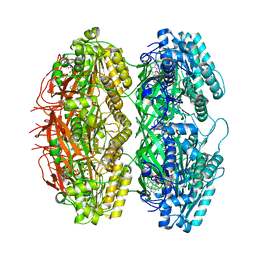 | | Structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
7X1Y
 
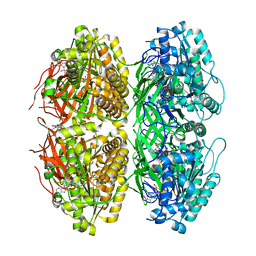 | | Structure of the phosphorylation-site double mutant S431A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
6EUM
 
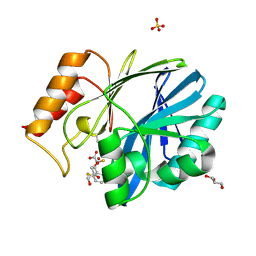 | | CRYSTAL STRUCTURE OF BCII METALLO-BETA-LACTAMASE IN COMPLEX WITH DZ-307 | | Descriptor: | (~{Z})-2-sulfanyl-3-[2,3,6-tris(fluoranyl)phenyl]prop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, El-Husseiny, A, Brem, J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
6F2N
 
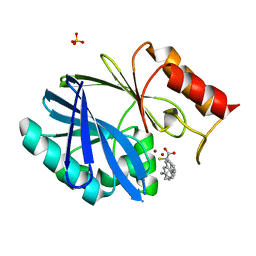 | | Crystal structure of BCII Metallo-beta-lactamase in complex with KDU197 | | Descriptor: | (~{Z})-3-[2-(naphthalen-2-ylmethyl)phenyl]-2-sulfanyl-prop-2-enoic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | McDonough, M.A, El-Hussein, A, Schofield, C.J, Zhang, D, Brem, J. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
6EWE
 
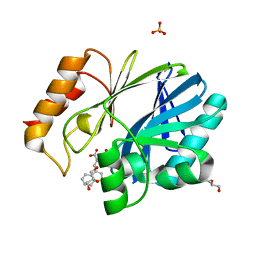 | | Crystal structure of BCII Metallo-beta-lactamase in complex with DZ-308 | | Descriptor: | (~{Z})-3-(1-benzothiophen-3-yl)-2-sulfanyl-prop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, El-Husseiny, A, Brem, J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
5JMX
 
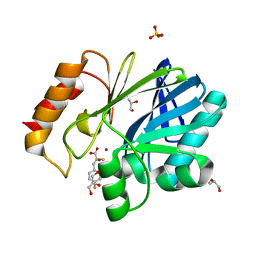 | | Crystal Structure of BcII metallo-beta-lactamase in complex with DZ-305 | | Descriptor: | (2Z)-3-(4-fluorophenyl)-2-sulfanylprop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, Brem, J. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
1KS5
 
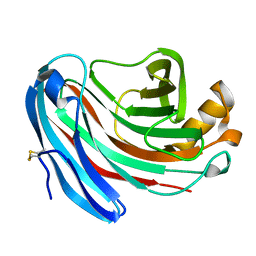 | | Structure of Aspergillus niger endoglucanase | | Descriptor: | Endoglucanase A | | Authors: | Khademi, S, Zhang, D, Swanson, S.M, Wartenberg, A, Witte, C, Meyer, E.F. | | Deposit date: | 2002-01-10 | | Release date: | 2003-01-21 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determination of the structure of an endoglucanase from Aspergillus niger and its mode of inhibition by palladium chloride.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6WZ8
 
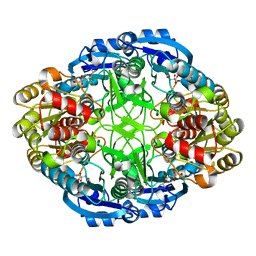 | | Complex of Pseudomonas 7A Glutaminase-Asparaginase with Citrate Anion at pH 5.5. Covalent Acyl-Enzyme Intermediate | | Descriptor: | CITRIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
6WYY
 
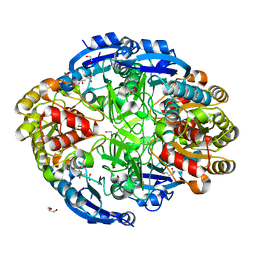 | | Crystal structure of Pseudomonas 7A Glutaminase-Asparaginase in complex with L-Glu at pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
6WZ4
 
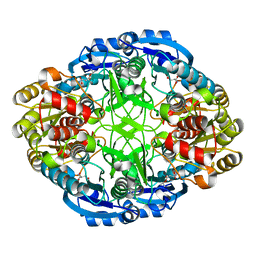 | | Complex of mutant (K173M) of Pseudomonas 7A Glutaminase-Asparaginase with L-Asp at pH 6. Covalent acyl-enzyme intermediate | | Descriptor: | ASPARTIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
1KS4
 
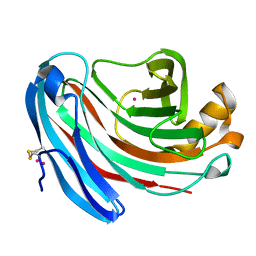 | | The structure of Aspergillus niger endoglucanase-palladium complex | | Descriptor: | Endoglucanase A, PALLADIUM ION | | Authors: | Khademi, S, Zhang, D, Swanson, S.M, Wartenberg, A, Witte, C, Meyer, E.F. | | Deposit date: | 2002-01-10 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the structure of an endoglucanase from Aspergillus niger and its mode of inhibition by palladium chloride.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6WYZ
 
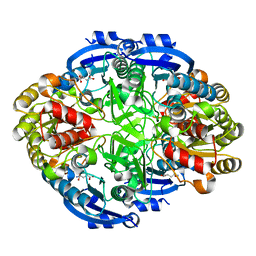 | | Crystal structure of Pseudomonas 7A Glutaminase-Asparaginase (mutant K173M) in complex with D-Glu at pH 5.5 | | Descriptor: | D-GLUTAMIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
5I9D
 
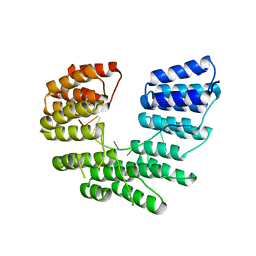 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8A2 in complex with its target RNA U8A2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*G*UP*UP*UP*UP*AP*AP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8A2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9H
 
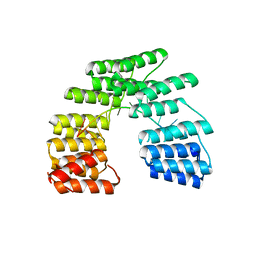 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8G2 in complex with its target RNA U8G2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*GP*GP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8G2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9G
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8C2 in complex with its target RNA U8C2 | | Descriptor: | RNA (5'-R(*GP*GP*G*GP*UP*UP*UP*UP*CP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8C2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9F
 
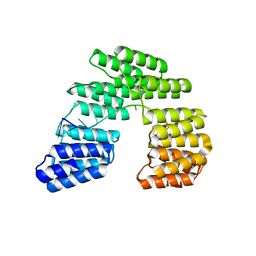 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U10 in complex with its target RNA U10 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U10 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, Y. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
7XA7
 
 | | Crystal structure of SARS-CoV-2 receptor-binding domain in complex with intermediate horseshoe bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Tang, L.F, Zhang, D, Han, P, Qi, J.X. | | Deposit date: | 2022-03-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of SARS-CoV-2 and its variants binding to intermediate horseshoe bat ACE2.
Int J Biol Sci, 18, 2022
|
|
6M1I
 
 | | CryoEM structure of human PAC1 receptor in complex with PACAP38 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
8P3X
 
 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Z
 
 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 2 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
