7YP9
 
 | |
2NML
 
 | | Crystal structure of HEF2/ERH at 1.55 A resolution | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Jin, T.C, Guo, F, Serebriiskii, I.G, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2006-10-21 | | Release date: | 2006-10-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55 A resolution X-ray crystal structure of HEF2/ERH and insights into its transcriptional and cell-cycle interaction networks.
Proteins, 68, 2007
|
|
7YPB
 
 | |
7YPA
 
 | |
6KCV
 
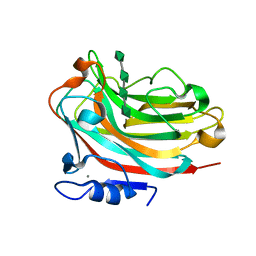 | | Structure of alginate lyase Aly36B mutant K143A/Y185A in complex with alginate tetrasaccharide | | Descriptor: | Alginate lyase, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Dong, F, Zhang, Y.Z, Chen, X.L. | | Deposit date: | 2019-06-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
J.Mol.Biol., 431, 2019
|
|
1ZCN
 
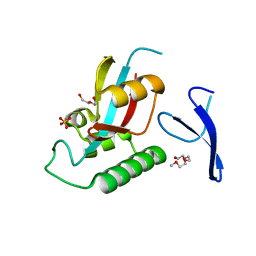 | | human Pin1 Ng mutant | | Descriptor: | PENTAETHYLENE GLYCOL, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Nguyen, H, Dendel, G, Bowman, M.E, Gruebele, M, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-04-12 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6LK8
 
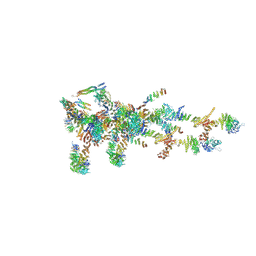 | | Structure of Xenopus laevis Cytoplasmic Ring subunit. | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Shi, Y, Huang, G, Yan, C, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis.
Cell Res., 30, 2020
|
|
1NW3
 
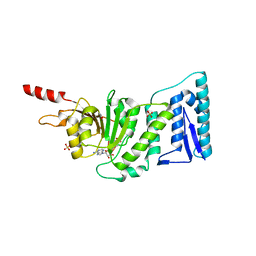 | | Structure of the Catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Min, J.R, Feng, Q, Li, Z.H, Zhang, Y, Xu, R.M. | | Deposit date: | 2003-02-05 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase
Cell(Cambridge,Mass.), 112, 2003
|
|
1P51
 
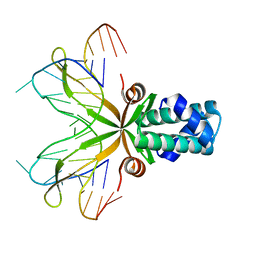 | | Anabaena HU-DNA cocrystal structure (AHU6) | | Descriptor: | 5'-D(*GP*CP*AP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*T)-3', DNA-binding protein HU | | Authors: | Swinger, K.K, Lemberg, K.M, Zhang, Y, Rice, P.A. | | Deposit date: | 2003-04-24 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexible DNA bending in HU-DNA cocrystal structures
Embo J., 22, 2003
|
|
1P3R
 
 | | Crystal structure of the phosphotyrosin binding domain(PTB) of mouse Disabled 1(Dab1) | | Descriptor: | Disabled homolog 2 | | Authors: | Yun, M, Keshvara, L, Park, C.G, Zhang, Y.M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.W. | | Deposit date: | 2003-04-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2.
J.Biol.Chem., 278, 2003
|
|
6IXJ
 
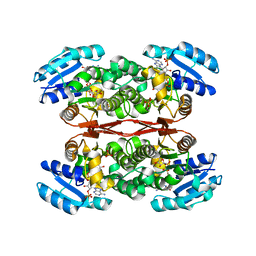 | | The crystal structure of sulfoacetaldehyde reductase from Klebsiella oxytoca | | Descriptor: | 2-hydroxyethylsulfonic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sulfoacetaldehyde reductase | | Authors: | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural investigation of sulfoacetaldehyde reductase fromKlebsiella oxytoca.
Biochem. J., 476, 2019
|
|
1F47
 
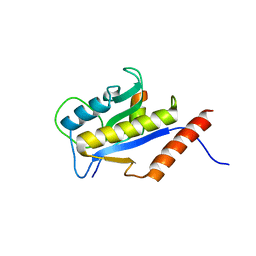 | | THE BACTERIAL CELL-DIVISION PROTEIN ZIPA AND ITS INTERACTION WITH AN FTSZ FRAGMENT REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CELL DIVISION PROTEIN FTSZ, CELL DIVISION PROTEIN ZIPA | | Authors: | Mosyak, L, Zhang, Y, Glasfeld, E, Stahl, M, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography.
EMBO J., 19, 2000
|
|
1Z68
 
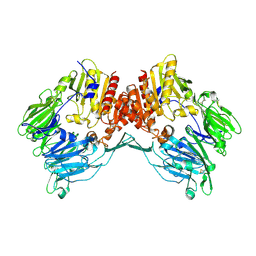 | | Crystal Structure Of Human Fibroblast Activation Protein alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aertgeerts, K, Levin, I, Shi, L, Prasad, G.S, Zhang, Y, Kraus, M.L, Salakian, S, Snell, G.P, Sridhar, V, Wijnands, R, Tennant, M.G. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha.
J.Biol.Chem., 280, 2005
|
|
2K29
 
 | |
1C24
 
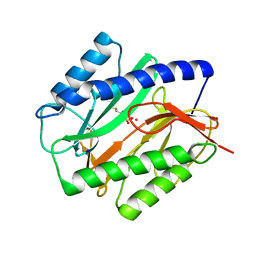 | | E. COLI METHIONINE AMINOPEPTIDASE: METHIONINE PHOSPHINATE COMPLEX | | Descriptor: | (1-AMINO-3-METHYLSULFANYL-PROPYL)-PHOSPHINIC ACID, COBALT (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Lowther, W.T, Zhang, Y, Sampson, P.B, Honek, J.F, Matthews, B.W. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues.
Biochemistry, 38, 1999
|
|
1ZYW
 
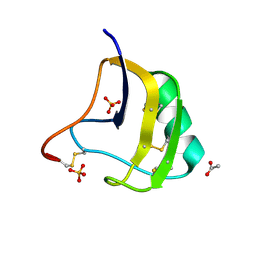 | | Crystal Structure Of Mutant K8DP9SR58KP60G Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | ACETATE ION, Alpha-like neurotoxin BmK-I, SULFATE ION | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-13 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1GN6
 
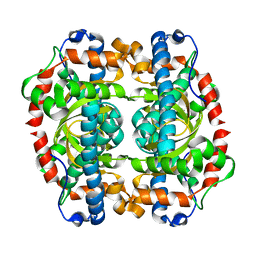 | | G152A mutant of Mycobacterium tuberculosis iron-superoxide dismutase. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Saward, S, Erskine, P.T, Badasso, M.O, Wood, S.P, Zhang, Y, Young, D.B. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-Ray Structure Analysis of an Engineered Fe-Superoxide Dismutase Gly-Ala Mutant with Significantly Reduced Stability to Denaturant
FEBS Lett., 387, 1996
|
|
7C1K
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant R148A | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7F65
 
 | | Bacetrial Cocaine Esterase with mutations T172R/G173Q/V116K/S117A/A51L, bound to benzoic acid | | Descriptor: | BENZOIC ACID, Cocaine esterase, SULFATE ION | | Authors: | Ouyang, P.F, Zhang, Y, Tong, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Computational Design and Crystal Structure of a Highly Efficient Benzoylecgonine Hydrolase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5Y3X
 
 | | Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis | | Descriptor: | Beta-xylanase | | Authors: | Liu, X, Sun, L.C, Zhang, Y.B, Liu, T.F, Xin, F.J. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Thermophilic Adaption Mechanism of Endo-1,4-beta-Xylanase from Caldicellulosiruptor owensensis.
J. Agric. Food Chem., 66, 2018
|
|
5U9D
 
 | | Discovery of a potent BTK inhibitor with a novel binding mode using parallel selections with a DNA-encoded chemical library | | Descriptor: | (R)-N-methyl-2-(3-((quinoxalin-6-ylamino)methyl)furan-2-carbonyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cuozzo, J.W, Centrella, P.A, Gikunju, D, Habeshian, S, Hupp, C.D, Keefe, A.D, Sigel, E, Soutter, H.H, Thomson, H.A, Zhang, Y, Clark, M.A. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a Potent BTK Inhibitor with a Novel Binding Mode by Using Parallel Selections with a DNA-Encoded Chemical Library.
Chembiochem, 18, 2017
|
|
4OHR
 
 | | Crystal structure of MilB from Streptomyces rimofaciens | | Descriptor: | CMP/hydroxymethyl CMP hydrolase | | Authors: | Zhao, G, Zhang, Y, Liu, G, Wu, G, He, X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry
Nucleic Acids Res., 42, 2014
|
|
5UIF
 
 | | Crystal Structure of Native Ps01740 | | Descriptor: | Ps01740 | | Authors: | LeVieux, J, Baas, B.J, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2017-01-13 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Kinetic and structural characterization of a cis-3-Chloroacrylic acid dehalogenase homologue in Pseudomonas sp. UW4: A potential step between subgroups in the tautomerase superfamily.
Arch. Biochem. Biophys., 636, 2017
|
|
7CKQ
 
 | |
3T0K
 
 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans with bound trisulfide and decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-20 | | Release date: | 2012-05-16 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
