1T8W
 
 | |
1T8S
 
 | |
1T8Y
 
 | |
1T8R
 
 | |
9ATN
 
 | |
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | 分子名称: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | 著者 | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | 登録日 | 2023-03-20 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | 分子名称: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | 著者 | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | 登録日 | 2023-03-20 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISI
 
 | | Photochromobilin-free form of Arabidopsis thaliana phytochrome A - apo-AtphyA | | 分子名称: | Phytochrome A | | 著者 | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | 登録日 | 2023-03-20 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
4S00
 
 | |
4RZZ
 
 | |
4RZY
 
 | |
4S01
 
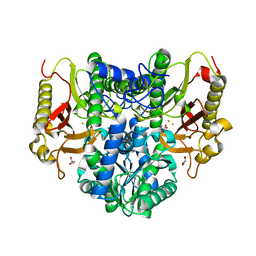 | |
9C11
 
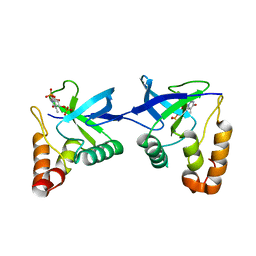 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36R at cryogenic temperature | | 分子名称: | CALCIUM ION, Nuclease A, THYMIDINE-3',5'-DIPHOSPHATE | | 著者 | Zhang, Y, Schlessman, J.L, Siegler, M.A, Garcia-Moreno E, B. | | 登録日 | 2024-05-28 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Domain-swapping promoted by the introduction of a charge in the hydrophobic interior of a protein
To Be Published
|
|
4RT7
 
 | | Crystal Structure of FLT3 with a small molecule inhibitor | | 分子名称: | 1-(5-tert-butyl-1,2-oxazol-3-yl)-3-(4-{7-[2-(morpholin-4-yl)ethoxy]imidazo[2,1-b][1,3]benzothiazol-2-yl}phenyl)urea, Receptor-type tyrosine-protein kinase FLT3 | | 著者 | Zhang, Y, Zhang, C. | | 登録日 | 2014-11-13 | | 公開日 | 2015-04-22 | | 最終更新日 | 2015-06-17 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Characterizing and Overriding the Structural Mechanism of the Quizartinib-Resistant FLT3 "Gatekeeper" F691L Mutation with PLX3397.
Cancer Discov, 5, 2015
|
|
6XNY
 
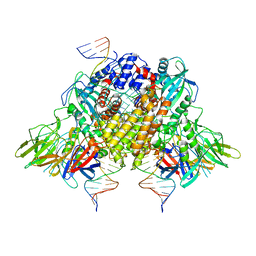 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Paired-Form) | | 分子名称: | 12RSS integration strand (55-mer), 12RSS signal DNA top strand (34-mer), 23RSS integration strand (66-mer), ... | | 著者 | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | 登録日 | 2020-07-05 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6XNX
 
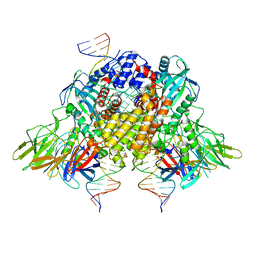 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Dynamic-Form) | | 分子名称: | 12RSS integration strand DNA (55-MER), 12RSS signal top strand DNA (34-MER), 23RSS integration strand DNA (66-MER), ... | | 著者 | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | 登録日 | 2020-07-05 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
4XVC
 
 | |
8AEY
 
 | | 3 A CRYO-EM STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS FERRITIN FROM TIMEPIX3 detector | | 分子名称: | Ferritin BfrB | | 著者 | Zhang, Y, van Schayck, J.P, Knoops, K, Peters, P.J, Ravelli, R.B.G. | | 登録日 | 2022-07-14 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Integration of an Event-driven Timepix3 Hybrid Pixel Detector into a Cryo-EM Workflow
Microsc Microanal, 2023
|
|
6XNZ
 
 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Target Capture Complex | | 分子名称: | 12RSS integration strand (34-mer), 12RSS non-integration strand (34-mer), 23RSS integration strand (45-mer), ... | | 著者 | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | 登録日 | 2020-07-05 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
4XV9
 
 | | B-Raf Kinase domain in complex with PLX5568 | | 分子名称: | N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}-4-(trifluoromethyl)benzenesulfonamide, SULFATE ION, Serine/threonine-protein kinase B-raf | | 著者 | zhang, Y, zhang, c, wang, w. | | 登録日 | 2015-01-26 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XZ6
 
 | | TmoX in complex with TMAO | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | 著者 | Zhang, Y.Z, Li, C.Y. | | 登録日 | 2015-02-04 | | 公開日 | 2015-08-19 | | 最終更新日 | 2015-10-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanistic Insight into Trimethylamine N-Oxide Recognition by the Marine Bacterium Ruegeria pomeroyi DSS-3
J.Bacteriol., 197, 2015
|
|
5GMR
 
 | |
5GMS
 
 | |
5GSN
 
 | | Tmm in complex with methimazole | | 分子名称: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, ... | | 著者 | Zhang, Y.Z, Li, C.Y. | | 登録日 | 2016-08-16 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.203 Å) | | 主引用文献 | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5LG4
 
 | |
