4O9F
 
 | |
8HI5
 
 | |
8HI4
 
 | |
8HI6
 
 | |
4JDL
 
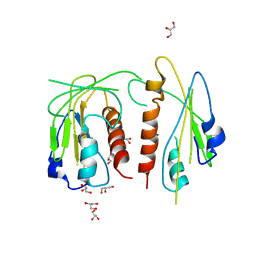 | |
5VJJ
 
 | | Crystal structure of the flax-rust effector AvrP | | 分子名称: | Avirulence protein AvrP123, ZINC ION | | 著者 | Zhang, X, Ericsson, D.J, Williams, S.J, Kobe, B. | | 登録日 | 2017-04-19 | | 公開日 | 2017-08-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Crystal structure of the Melampsora lini effector AvrP reveals insights into a possible nuclear function and recognition by the flax disease resistance protein P.
Mol. Plant Pathol., 19, 2018
|
|
5TEC
 
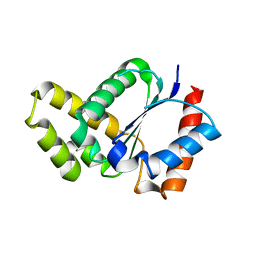 | | Crystal structure of the TIR domain from the Arabidopsis thaliana NLR protein SNC1 | | 分子名称: | Protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 | | 著者 | Zhang, X, Bentham, A, Ve, T, Williams, S.J, Kobe, B. | | 登録日 | 2016-09-20 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4OA7
 
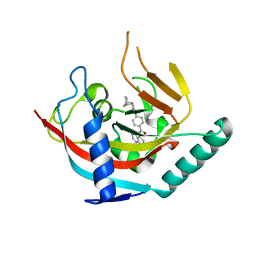 | | Crystal structure of Tankyrase1 in complex with IWR1 | | 分子名称: | 4-[(3aR,4R,7S,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | 著者 | Zhang, X, He, H. | | 登録日 | 2014-01-03 | | 公開日 | 2015-01-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
5K3H
 
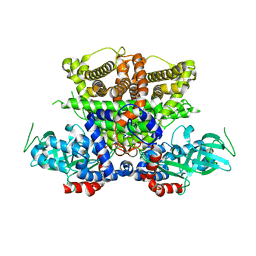 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-II | | 分子名称: | Acyl-coenzyme A oxidase | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K3G
 
 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-I | | 分子名称: | Acyl-coenzyme A oxidase | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.859 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3IYL
 
 | | Atomic CryoEM Structure of a Nonenveloped Virus Suggests How Membrane Penetration Protein is Primed for Cell Entry | | 分子名称: | Core protein VP6, MYRISTIC ACID, Outer capsid VP4, ... | | 著者 | Zhang, X, Jin, L, Fang, Q, Hui, W, Zhou, Z.H. | | 登録日 | 2010-02-02 | | 公開日 | 2010-05-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry.
Cell(Cambridge,Mass.), 141, 2010
|
|
5K3J
 
 | | Crystals structure of Acyl-CoA oxidase-2 in Caenorhabditis elegans bound with FAD, ascaroside-CoA, and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3J4U
 
 | | A new topology of the HK97-like fold revealed in Bordetella bacteriophage: non-covalent chainmail secured by jellyrolls | | 分子名称: | cementing protein, major capsid protein | | 著者 | Zhang, X, Guo, H, Jin, L, Czornyj, E, Hodes, A, Hui, W.H, Nieh, A.W, Miller, J.F, Zhou, Z.H. | | 登録日 | 2013-10-09 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution.
Elife, 2, 2013
|
|
5K3I
 
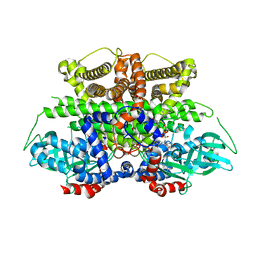 | | Crystal structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans complexed with FAD and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.683 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4JDA
 
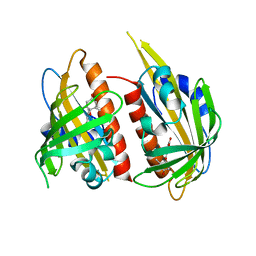 | | Complex structure of abscisic acid receptor PYL3 with (-)-ABA | | 分子名称: | (2Z,4E)-5-[(1R)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | 著者 | Zhang, X, Wang, G, Chen, Z. | | 登録日 | 2013-02-24 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
2MX6
 
 | |
1DYB
 
 | |
2P0P
 
 | |
1DYE
 
 | |
1DYA
 
 | |
1DYD
 
 | |
1DYC
 
 | |
2RFD
 
 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | 分子名称: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor, SULFATE ION | | 著者 | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | 登録日 | 2007-09-28 | | 公開日 | 2007-12-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2P0Q
 
 | |
2RF9
 
 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | 分子名称: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | 著者 | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | 登録日 | 2007-09-28 | | 公開日 | 2007-12-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
