5CG8
 
 | | NgTET1 in complex with 5hmC DNA | | Descriptor: | 2-OXOGLUTARIC ACID, DNA (5'-D(*AP*GP*AP*AP*TP*TP*CP*CP*GP*TP*TP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*(5HC)P*GP*GP*AP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
4EW0
 
 | | mouse MBD4 glycosylase domain in complex with a G:5hmU (5-hydroxymethyluracil) mismatch | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*(5HU)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
6O0W
 
 | | Crystal structure of the TIR domain from the grapevine disease resistance protein RUN1 in complex with NADP+ and Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-2'-5'-DIPHOSPHATE, TIR-NB-LRR type resistance protein RUN1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
1RKV
 
 | | Structure of Phosphate complex of ThrH from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Singh, S.K, Yang, K, Subramanian, K, Karthikeyan, S, Huynh, T, Zhang, X, Phillips, M.A, Zhang, H. | | Deposit date: | 2003-11-23 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The thrH Gene Product of Pseudomonas aeruginosa Is a Dual Activity Enzyme with a Novel Phosphoserine:Homoserine Phosphotransferase Activity.
J.Biol.Chem., 279, 2004
|
|
6O0V
 
 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
8JEQ
 
 | | Crystal structure of Tiragolumab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEP
 
 | | Crystal structure of Ociperlimab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEO
 
 | | Crystal structure of TIGIT in complexed with Tiragolumab | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
6O1B
 
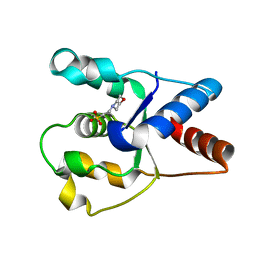 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
7LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
8BQN
 
 | |
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
4EVV
 
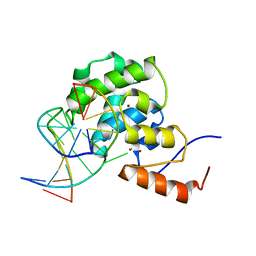 | | mouse MBD4 glycosylase domain in complex with a G:T mismatch | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*TP*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4AU6
 
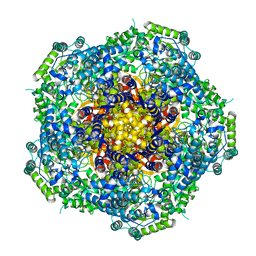 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Location of the Dsrna-Dependent Polymerase, Vp1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
4EW4
 
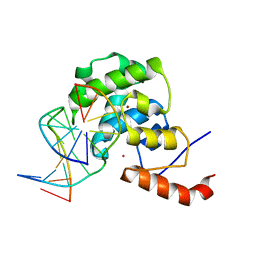 | | mouse MBD4 glycosylase domain in complex with DNA containing a ribose sugar | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*GP*(3DR)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4, ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4F0P
 
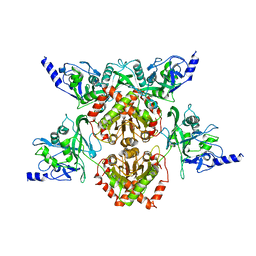 | | MspJI Restriction Endonuclease - P31 Form | | Descriptor: | MAGNESIUM ION, Restriction endonuclease | | Authors: | Horton, J.R, Mabuchi, M, Cohen-Karni, D, Zhang, X, Griggs, R, Samaranayake, M, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2012-05-04 | | Release date: | 2012-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
4F0Q
 
 | | MspJI Restriction Endonuclease - P21 Form | | Descriptor: | MAGNESIUM ION, Restriction endonuclease | | Authors: | Horton, J.R, Mabuchi, M, Cohen-Karni, D, Zhang, X, Griggs, R, Samaranayake, M, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2012-05-04 | | Release date: | 2012-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
1F4H
 
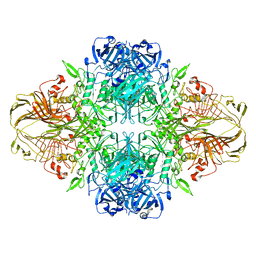 | | E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1F4A
 
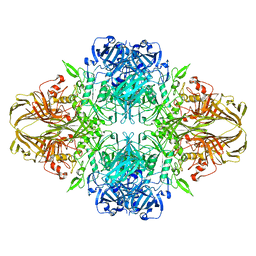 | | E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1F6W
 
 | |
4G3H
 
 | | Crystal structure of helicobacter pylori arginase | | Descriptor: | Arginase (RocF), MANGANESE (II) ION | | Authors: | Zhang, J, Zhang, X, Li, D, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function studies on Helicobacter pylori arginase
To be Published
|
|
5XNO
 
 | | Structure of M-LHCII and CP24 complexes in the unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
1EUE
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oganesyan, V, Zhang, X. | | Deposit date: | 2000-04-19 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of redox potential in electron transfer proteins: effects of complex formation on the active site microenvironment of cytochrome b5.
FARADAY DISC.CHEM.SOC, 116, 2001
|
|
