5TEC
 
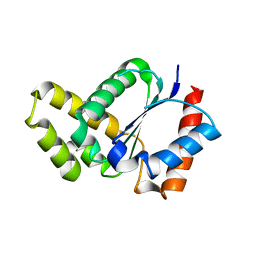 | | Crystal structure of the TIR domain from the Arabidopsis thaliana NLR protein SNC1 | | 分子名称: | Protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 | | 著者 | Zhang, X, Bentham, A, Ve, T, Williams, S.J, Kobe, B. | | 登録日 | 2016-09-20 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5K3J
 
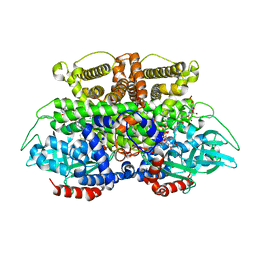 | | Crystals structure of Acyl-CoA oxidase-2 in Caenorhabditis elegans bound with FAD, ascaroside-CoA, and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
191L
 
 | |
3IYK
 
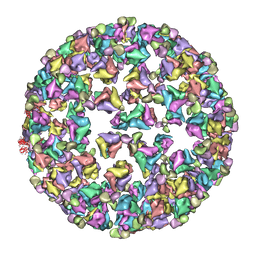 | | Bluetongue virus structure reveals a sialic acid binding domain, amphipathic helices and a central coiled coil in the outer capsid proteins | | 分子名称: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, VP2, VP5 | | 著者 | Zhang, X, Boyce, M, Bhattacharya, B, Zhang, X, Schein, S, Roy, P, Zhou, Z.H. | | 登録日 | 2010-01-25 | | 公開日 | 2010-04-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Bluetongue virus coat protein VP2 contains sialic acid-binding domains, and VP5 resembles enveloped virus fusion proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7CBQ
 
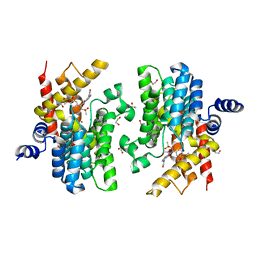 | | Crystal structure of PDE4D catalytic domain in complex with Apremilast | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-{2-[(1S)-1-(3-ethoxy-4-methoxyphenyl)-2-(methylsulfonyl)ethyl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}acetamide, ... | | 著者 | Zhang, X.L, Xu, Y.C. | | 登録日 | 2020-06-13 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | 分子名称: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | 著者 | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | 登録日 | 2010-08-23 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
5XBM
 
 | | Structure of SCARB2-JL2 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | 著者 | Zhang, X, Yang, P, Wang, N, Zhang, J, Li, J, Guo, H, Yin, X, Rao, Z, Wang, X, Zhang, L. | | 登録日 | 2017-03-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.501 Å) | | 主引用文献 | The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection.
Protein Cell, 8, 2017
|
|
5EBC
 
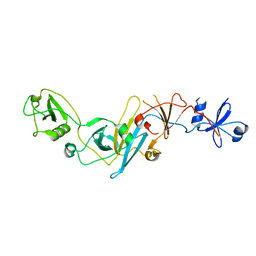 | | Crystal structure of EccB1 of Mycobacterium tuberculosis in spacegroup P21 (state III) | | 分子名称: | CALCIUM ION, ESX-1 secretion system protein eccB1 | | 著者 | Zhang, X.L, Qi, C, Xie, X.Q, Li, D.F, Bi, L.J. | | 登録日 | 2015-10-19 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystallographic observation of the movement of the membrane-distal domain of the T7SS core component EccB1 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1CDZ
 
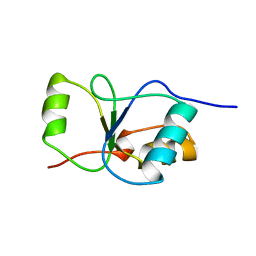 | | BRCT DOMAIN FROM DNA-REPAIR PROTEIN XRCC1 | | 分子名称: | PROTEIN (DNA-REPAIR PROTEIN XRCC1) | | 著者 | Zhang, X, Morera, S, Bates, P, Whitehead, P, Coffer, A, Hainbucher, K, Nash, R, Sternberg, M, Lindahl, T, Freemont, P. | | 登録日 | 1999-03-04 | | 公開日 | 2000-02-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of an XRCC1 BRCT domain: a new protein-protein interaction module.
EMBO J., 17, 1998
|
|
192L
 
 | |
7CBJ
 
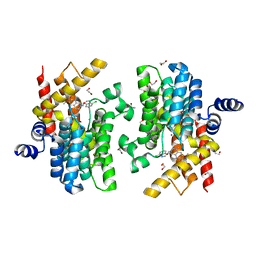 | | Crystal structure of PDE4D catalytic domain in complex with compound 36 | | 分子名称: | (1S)-1-[(7-chloranyl-1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydro-1H-isoquinoline-2-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | 著者 | Zhang, X.L, Xu, Y.C. | | 登録日 | 2020-06-12 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
190L
 
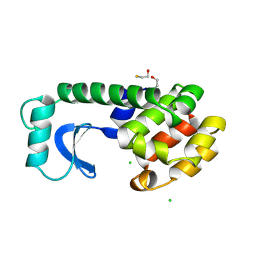 | |
4KSY
 
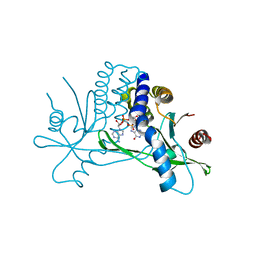 | |
5EBD
 
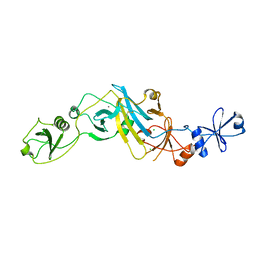 | | Crystal structure of EccB1 of Mycobacterium tuberculosis in spacegroup P21 (state IV) | | 分子名称: | CALCIUM ION, CHLORIDE ION, ESX-1 secretion system protein eccB1 | | 著者 | Zhang, X.L, Qi, C, Xie, X.Q, Li, D.F, Bi, L.J. | | 登録日 | 2015-10-19 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystallographic observation of the movement of the membrane-distal domain of the T7SS core component EccB1 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1DYB
 
 | |
1DYE
 
 | |
1DYA
 
 | |
1DYD
 
 | |
1DYC
 
 | |
2CSE
 
 | | Features of Reovirus Outer-Capsid Protein mu1 Revealed by Electron and Image Reconstruction of the virion at 7.0-A Resolution | | 分子名称: | Minor core protein lambda 3, Sigma 2 protein, guanylyltransferase, ... | | 著者 | Zhang, X, Ji, Y, Zhang, L, Harrison, S.C, Marinescu, D.C, Nibert, M.L, Baker, T.S. | | 登録日 | 2005-05-21 | | 公開日 | 2005-10-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution.
Structure, 13, 2005
|
|
8YE5
 
 | |
8JSD
 
 | |
6J4M
 
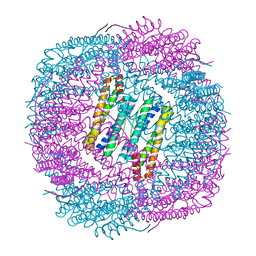 | | Thermal treated soybean seed H-2 ferritin | | 分子名称: | Ferritin, MAGNESIUM ION | | 著者 | Zhang, X, Zang, J, Chen, H, Zhou, K, Zhao, G. | | 登録日 | 2019-01-09 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Thermostability of protein nanocages: the effect of natural extra peptide on the exterior surface.
Rsc Adv, 9, 2019
|
|
3OQU
 
 | | Crystal structure of native abscisic acid receptor PYL9 with ABA | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL9 | | 著者 | Zhang, X, Zhang, Q, Chen, Z. | | 登録日 | 2010-09-04 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | 著者 | Zhang, X, Chen, Z. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
