8IFL
 
 | |
8ISM
 
 | | HSA-Pt compound complex | | Descriptor: | 7-(2-azanyl-5-chloranyl-phenyl)-3$l^{3}-thia-5,6$l^{4}-diaza-2$l^{3}-platinatricyclo[6.4.0.0^{2,6}]dodeca-1(12),3,6,8,10-pentaen-4-amine, PALMITIC ACID, Serum albumin | | Authors: | Zhang, J.Z, Zhang, Z.L. | | Deposit date: | 2023-03-21 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | HSA-Pt compound complex
To Be Published
|
|
2NWM
 
 | |
3STA
 
 | | Crystal structure of ClpP in tetradecameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
3ST9
 
 | | Crystal structure of ClpP in heptameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
8K1I
 
 | |
5X5R
 
 | |
8K34
 
 | | Cryo-EM structure of SPARTA gRNA binary complex | | Descriptor: | MAGNESIUM ION, Piwi domain-containing protein, RNA (5'-R(P*AP*AP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3'), ... | | Authors: | Zhang, J.T, Jia, N. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Target ssDNA activates the NADase activity of prokaryotic SPARTA immune system.
Nat.Chem.Biol., 20, 2024
|
|
7Y85
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease bound to self RNA target | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y81
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA complex bound to non-self RNA target | | Descriptor: | MAGNESIUM ION, Non-self RNA target, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y80
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA binary complex | | Descriptor: | MAGNESIUM ION, RAMP superfamily protein, ZINC ION, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y84
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y82
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA complex bound to self RNA target | | Descriptor: | MAGNESIUM ION, RAMP superfamily protein, Self RNA target, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y83
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease bound to non-self RNA target | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
5X5M
 
 | |
4LCK
 
 | |
7DGD
 
 | | apo state of class C GPCR | | Descriptor: | Metabotropic glutamate receptor 1 | | Authors: | Zhang, J.Y, Wu, L.J, Luo, F, Hua, T, Liu, Z.J. | | Deposit date: | 2020-11-11 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural insights into the activation initiation of full-length mGlu1.
Protein Cell, 12, 2021
|
|
7DGE
 
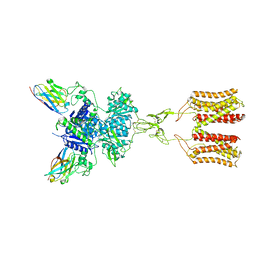 | | intermediate state of class C GPCR | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 1, nanobody | | Authors: | Zhang, J.Y, Wu, L.J, Luo, F, Hua, T, Liu, Z.J. | | Deposit date: | 2020-11-11 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural insights into the activation initiation of full-length mGlu1.
Protein Cell, 12, 2021
|
|
5H3V
 
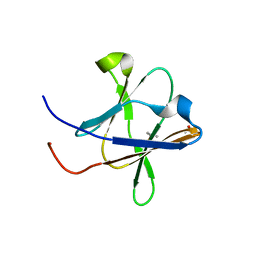 | | Crystal structure of a Type IV Secretion System Component CagX in Helicobacter pylori | | Descriptor: | Cag8, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL | | Authors: | Zhang, J, Wu, Y, Zhao, Y, Sun, L, Keegan, R.M, Liu, Y, Isupov, M.N. | | Deposit date: | 2016-10-27 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the type IV secretion system component CagX from Helicobacter pylori
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
8IW5
 
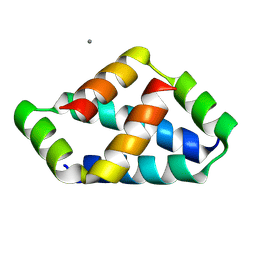 | | Crystal structure of liprin-beta H2H3 dimer | | Descriptor: | CALCIUM ION, Liprin-beta-1 | | Authors: | Zhang, J, Chen, S, Wei, Z. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
8IW0
 
 | | Crystal structure of the KANK1/liprin-beta1 complex | | Descriptor: | Liprin-beta-1,KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Zhang, J, Chen, S, Wei, Z, Yu, C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
7DDE
 
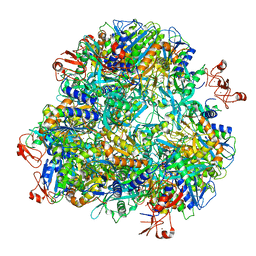 | | Cryo-EM structure of the Ape4 and Nbr1 complex | | Descriptor: | Aspartyl aminopeptidase 1,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
7DD9
 
 | | Cryo-EM structure of the Ams1 and Nbr1 complex | | Descriptor: | Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
6AR6
 
 | |
5ZY9
 
 | | Structural basis for a tRNA synthetase | | Descriptor: | (1R,2R)-2-[(2S,4E,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadeca-4,6-dien-2-yl]cyclopentanecarboxylic acid, Threonyl-tRNA synthase, ZINC ION | | Authors: | Zhang, J. | | Deposit date: | 2018-05-23 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for inhibition of
translational functions on a tRNA synthetase
To Be Published
|
|
