6LJ7
 
 | |
6LJ6
 
 | |
6LL9
 
 | |
1J55
 
 | | The Crystal Structure of Ca+-bound Human S100P Determined at 2.0A Resolution by X-ray | | Descriptor: | CALCIUM ION, S-100P PROTEIN | | Authors: | Zhang, H, Wang, G, Ding, Y, Wang, Z, Barraclough, R, Rudland, P.S, Fernig, D.G, Rao, Z. | | Deposit date: | 2002-01-25 | | Release date: | 2003-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure at 2A Resolution of the Ca2+-binding Protein S100P
J.Mol.Biol., 325, 2003
|
|
8K2E
 
 | | Crystal structure of YTHDC1 and Y3 complex | | Descriptor: | 2-chloranyl-6-(4-fluoranylphenoxy)benzoic acid, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Zhang, H.L, Yang, S.Y. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PocketFlow is a data-and-knowledge-driven structure-based molecular generative model
Nat. Mach. Intell., 6, 2024
|
|
4RT4
 
 | | Crystal structure of Dpy30 complexed with Bre2 | | Descriptor: | Peptide from COMPASS component BRE2, Protein dpy-30 homolog | | Authors: | Zhang, H.M, Li, M, Chang, W.R. | | Deposit date: | 2014-11-12 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural implications of Dpy30 oligomerization for MLL/SET1 COMPASS H3K4 trimethylation
Protein Cell, 6, 2015
|
|
7XB6
 
 | |
8J9A
 
 | |
7X7S
 
 | | Solution structure of human adenylate kinase 1 (hAK1) | | Descriptor: | Adenylate kinase isoenzyme 1 | | Authors: | Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ADP-Induced Conformational Transition of Human Adenylate Kinase 1 Is Triggered by Suppressing Internal Motion of alpha 3 alpha 4 and alpha 7 alpha 8 Fragments on the ps-ns Timescale.
Biomolecules, 12, 2022
|
|
6XP5
 
 | | Head-Middle module of Mediator | | Descriptor: | HEAT, Med22, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Zhang, H.Q, Chen, D.C, Kornberg, R.D. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
4RTA
 
 | |
8GS4
 
 | | Cryo-EM structure of human Neuroligin 2 | | Descriptor: | Neuroligin-2 | | Authors: | Zhang, H, Zhang, Z, Hou, M. | | Deposit date: | 2022-09-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Expression and structural analysis of human neuroligin 2 and neuroligin 3 implicated in autism spectrum disorders.
Front Endocrinol, 13, 2022
|
|
8KCB
 
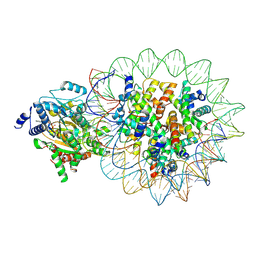 | | Complex of DDM1-nucleosome(H2A) complex with DDM1 bound to SHL2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Zhang, H, Zhang, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Mechanism of heterochromatin remodeling revealed by the DDM1 bound nucleosome structures.
Structure, 32, 2024
|
|
8KCC
 
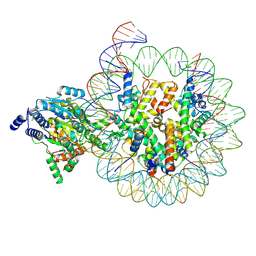 | | Complex of DDM1-nucleosome(H2A.W) complex with DDM1 bound to SHL2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Zhang, H, Zhang, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of heterochromatin remodeling revealed by the DDM1 bound nucleosome structures.
Structure, 32, 2024
|
|
8K98
 
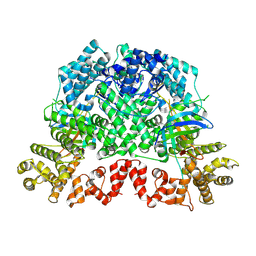 | | Cryo-EM structure of DSR2-TTP | | Descriptor: | a protein | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
8K9A
 
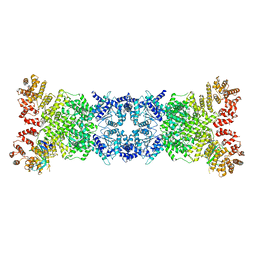 | | Cryo-EM structure of DSR2-DSAD1 state 2 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
5E09
 
 | |
5NBG
 
 | |
5E0C
 
 | |
6OAW
 
 | | Crystal structure of a CRISPR Cas-related protein | | Descriptor: | UNKNOWN ATOM OR ION, WYL1 | | Authors: | Zhang, H, Dong, C, Li, L, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-18 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the modulatory role of the accessory protein WYL1 in the Type VI-D CRISPR-Cas system.
Nucleic Acids Res., 47, 2019
|
|
6OYC
 
 | |
5GS5
 
 | | Crystal structure of apo rat STING | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of apo ratSTING
To Be Published
|
|
5H21
 
 | | Trimethoxy-ring inhibitor in complex with the first bromodomain of BRD4 | | Descriptor: | 3,4,5-trimethoxy-~{N}-(2-thiophen-2-ylethyl)benzamide, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of novel trimethoxy-ring BRD4 bromodomain inhibitors: AlphaScreen assay, crystallography and cell-based assay.
Medchemcomm, 8, 2017
|
|
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
1C1J
 
 | | STRUCTURE OF CADMIUM-SUBSTITUTED PHOSPHOLIPASE A2 FROM AGKISTRONDON HALYS PALLAS AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | BASIC PHOSPHOLIPASE A2, CADMIUM ION, octyl beta-D-glucopyranoside | | Authors: | Zhang, H.-l, Zhang, Y.-q, Song, S.-y, Zhou, Y, Lin, Z.-j. | | Deposit date: | 1999-07-22 | | Release date: | 2002-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Cadmium-substituted Phospholipase A2 from Agkistrodon halys
Pallas at 2.8 Angstroms Resolution
Protein Pept.Lett., 6, 1999
|
|
