7KJ5
 
 | | SARS-CoV-2 Spike Glycoprotein, prefusion with one RBD up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KJ2
 
 | | SARS-CoV-2 Spike Glycoprotein with one ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KJ4
 
 | | SARS-CoV-2 Spike Glycoprotein with three ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8XG2
 
 | | The structure of HLA-A/Pep14 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XES
 
 | | The structure of HLA-A/L1-1 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKE
 
 | | The structure of HLA-A/14-3-D | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | Authors: | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XFZ
 
 | | The structure of HLA-A/L1-2 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
6AR6
 
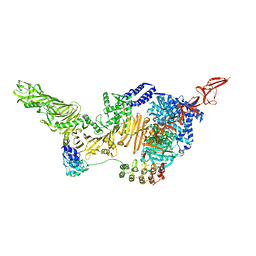 | |
4G42
 
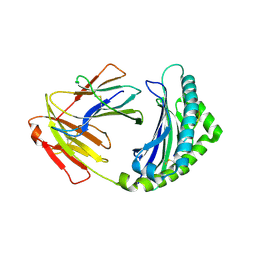 | | Structure of the Chicken MHC Class I Molecule BF2*0401 complexed to pepitde P8D | | Descriptor: | 8-MERIC PEPTIDE P8D, Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
4G43
 
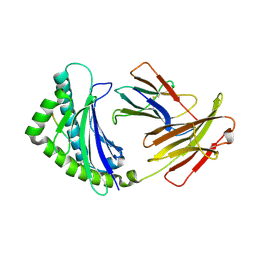 | | Structure of the chicken MHC class I molecule BF2*0401 complexed to P5E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-MERIC PEPTIDE P5E, Beta-2 microglobulin, ... | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
6BWF
 
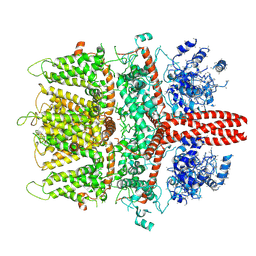 | | 4.1 angstrom Mg2+-unbound structure of mouse TRPM7 | | Descriptor: | TRPM7 | | Authors: | Zhang, J, Li, Z, Duan, J, Abiria, S.A, Clapham, D.E. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-15 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the mammalian TRPM7, a magnesium channel required during embryonic development.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2GMJ
 
 | | Structure of Porcine Electron Transfer Flavoprotein-Ubiquinone Oxidoreductase | | Descriptor: | Electron transfer flavoprotein-ubiquinone oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Zhang, J, Frerman, F.E, Kim, J.-J.P. | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8ISM
 
 | | HSA-Pt compound complex | | Descriptor: | 7-(2-azanyl-5-chloranyl-phenyl)-3$l^{3}-thia-5,6$l^{4}-diaza-2$l^{3}-platinatricyclo[6.4.0.0^{2,6}]dodeca-1(12),3,6,8,10-pentaen-4-amine, PALMITIC ACID, Serum albumin | | Authors: | Zhang, J.Z, Zhang, Z.L. | | Deposit date: | 2023-03-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | HSA-Pt compound complex
To Be Published
|
|
8W8D
 
 | |
7YUK
 
 | | Complex structure of BANP BEN domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*CP*GP*CP*GP*AP*GP*AP*G)-3'), GLYCEROL, Protein BANP | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7YUG
 
 | | Structure of human BANP BEN domain | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
2GMH
 
 | | Structure of Porcine Electron Transfer Flavoprotein-Ubiquinone Oxidoreductase in Complexed with Ubiquinone | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, Electron transfer flavoprotein-ubiquinone oxidoreductase, ... | | Authors: | Zhang, J, Frerman, F.E, Kim, J.-J.P. | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8T5U
 
 | | ATP-1 state of Bcs1 (C7 symmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-06-14 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8T14
 
 | | ADP-bound Bcs1 (C7 symmetrized) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8K34
 
 | | Cryo-EM structure of SPARTA gRNA binary complex | | Descriptor: | MAGNESIUM ION, Piwi domain-containing protein, RNA (5'-R(P*AP*AP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3'), ... | | Authors: | Zhang, J.T, Jia, N. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Target ssDNA activates the NADase activity of prokaryotic SPARTA immune system.
Nat.Chem.Biol., 20, 2024
|
|
7KRR
 
 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
7KRS
 
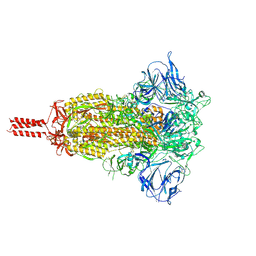 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
7KRQ
 
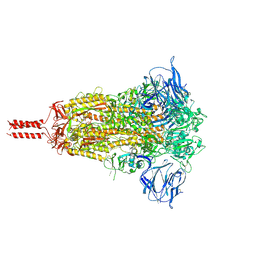 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
8TPL
 
 | | ATP-2 state of Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
