7O4Z
 
 | | Crystal structure of the carbonic anhydrase-like domain of CcmM from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | CHLORIDE ION, Carboxysome assembly protein CcmM, NICKEL (II) ION | | Authors: | Zang, K, Wang, H, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2021-04-07 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Scaffolding protein CcmM directs multiprotein phase separation in beta-carboxysome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7O54
 
 | | Crystal structure of the carbonic anhydrase-like domain of CcmM in complex with the C-terminal 17 residues of CcaA from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | CHLORIDE ION, Carbonic anhydrase, Carboxysome assembly protein CcmM, ... | | Authors: | Zang, K, Wang, H, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2021-04-07 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Scaffolding protein CcmM directs multiprotein phase separation in beta-carboxysome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2AP7
 
 | |
2AP8
 
 | |
2JTW
 
 | |
1DFS
 
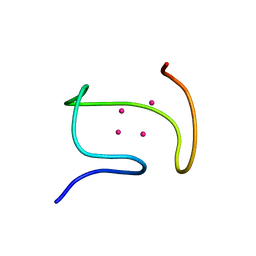 | | SOLUTION STRUCTURE OF THE ALPHA-DOMAIN OF MOUSE METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Zangger, K, Oz, G, Otvos, J.D, Armitage, I.M. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy.
Protein Sci., 8, 1999
|
|
1DFT
 
 | | SOLUTION STRUCTURE OF THE BETA-DOMAIN OF MOUSE METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Zangger, K, Oz, G, Otvos, J.D, Armitage, I.M. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy.
Protein Sci., 8, 1999
|
|
2KMT
 
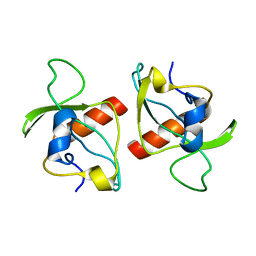 | |
2KV5
 
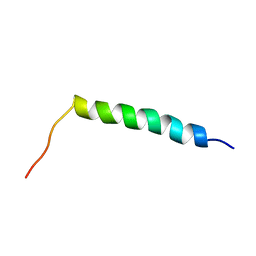 | |
6HH0
 
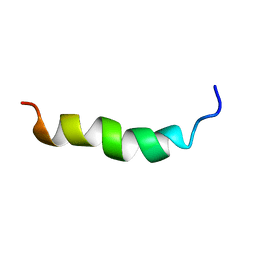 | |
6K7Q
 
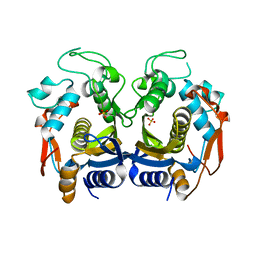 | | Crystal structure of thymidylate synthase from shrimp | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Ma, Q, Zang, K, Liu, C. | | Deposit date: | 2019-06-08 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural analysis of a shrimp thymidylate synthase reveals species-specific interactions with dUMP and raltitrexed.
J Oceanol Limnol, 38, 2020
|
|
5Y5P
 
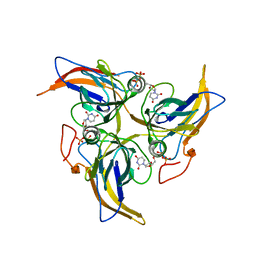 | |
5Y5Q
 
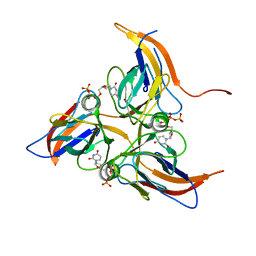 | |
5Y5O
 
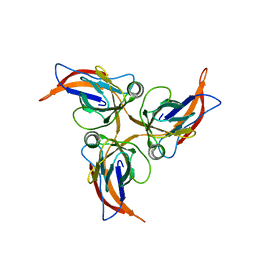 | |
7C4A
 
 | | nicA2 with cofactor FAD | | Descriptor: | Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xu, P, Zang, K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Deceleration Regulates Toxicant Release to Prevent Cell Damage in Pseudomonas putida S16 (DSM 28022).
Mbio, 11, 2020
|
|
7XYJ
 
 | | Structure of WSSV thymidylate synthase in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Ma, Q, Liu, C, Zang, K. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Structure of WSSV thymidylate synthase in complex with dUMP
To Be Published
|
|
7XYK
 
 | |
6K7R
 
 | | Crystal structure of thymidylate synthase from shrimp | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Ma, Q, Zang, K, Liu, C. | | Deposit date: | 2019-06-08 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural analysis of a shrimp thymidylate synthase reveals species-specific interactions with dUMP and raltitrexed.
J Oceanol Limnol, 38, 2020
|
|
6K7S
 
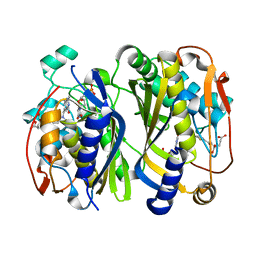 | | Crystal structure of thymidylate synthase from shrimp | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, TOMUDEX, Thymidylate synthase | | Authors: | Ma, Q, Zang, K, Liu, C. | | Deposit date: | 2019-06-08 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural analysis of a shrimp thymidylate synthase reveals species-specific interactions with dUMP and raltitrexed.
J Oceanol Limnol, 38, 2020
|
|
7NN6
 
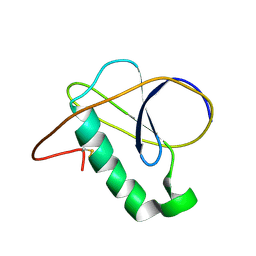 | |
8OZZ
 
 | |
5A4K
 
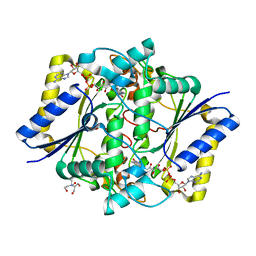 | | Crystal structure of the R139W variant of human NAD(P)H:quinone oxidoreductase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Strandback, E, Gudipati, V, Uhl, M.K, Rantase, D.M, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Catalytic competence, structure and stability of the cancer-associated R139W variant of the human NAD(P)H:quinone oxidoreductase 1 (NQO1).
FEBS J., 284, 2017
|
|
4CF6
 
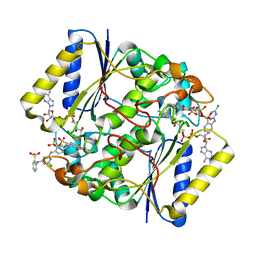 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with Cibacron blue at 2.7 A resolution | | Descriptor: | CIBACRON BLUE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
8ALO
 
 | | Heterodimer formation of sensory domains of Vibrio cholerae regulators ToxR and ToxS | | Descriptor: | Cholera toxin transcriptional activator, Transmembrane regulatory protein ToxS | | Authors: | Gubensaek, N, Sagmeister, T, Pavkov-Keller, T, Zangger, K, Buhlheller, C, Wagner, G.E. | | Deposit date: | 2022-08-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Vibrio cholerae's ToxRS bile sensing system.
Elife, 12, 2023
|
|
4CET
 
 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with dicoumarol at 2.2 A resolution | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
