4BOL
 
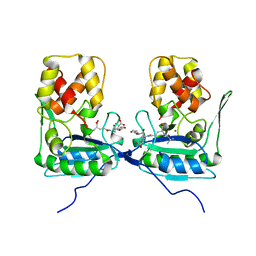 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with pentapeptide | | Descriptor: | AMPDH2, D-alanyl-N-[(2S,6R)-6-amino-6-carboxy-1-{[(1R)-1-carboxyethyl]amino}-1-oxohexan-2-yl]-D-glutamine, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E.E, Lastochkin, E, Zhang, W, Hellman, L.M, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-21 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
7AA8
 
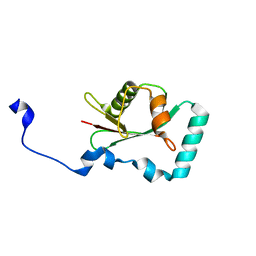 | | Structure of SCOC LIR bound to GABARAP | | Descriptor: | Chimera made of SCOC (6-23) + linker (GS) + GABARAP,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
7AA7
 
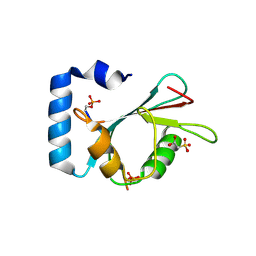 | | Structure of SCOC pS12/pS18 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pS12/pS18 SCOC LIR, sulfoacetic acid | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
6J1U
 
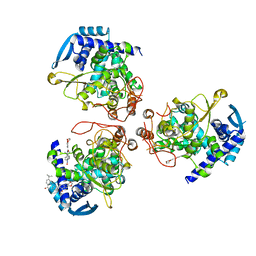 | | influenza virus nucleoprotein with a specific inhibitor | | Descriptor: | Nucleoprotein, ~{N}-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]-2,3-dihydro-1,4-benzodioxine-6-carboxamide | | Authors: | Pang, B, Zhang, W.Z, Zhang, H.M, Hao, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Novel Specific Inhibitor Targeting Influenza A Virus Nucleoprotein with Pleiotropic Inhibitory Effects on Various Steps of the Viral Life Cycle.
J.Virol., 95, 2021
|
|
8IDE
 
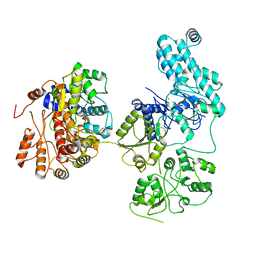 | | Structure of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme (TsaN) | | Descriptor: | MANGANESE (II) ION, N(6)-L-threonylcarbamoyladenine synthase | | Authors: | Zhang, Z.L, Jin, M.Q, Yu, Z.J, Chen, W, Wang, X.L, Lei, D.S, Zhang, W.H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure-function analysis of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme reveals a prototype of tRNA t6A and ct6A synthetases.
Nucleic Acids Res., 51, 2023
|
|
8WX7
 
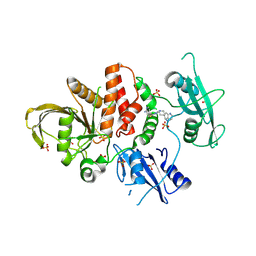 | | Crystal structure of SHP2 in complex with JAB-3186 | | Descriptor: | (5~{S})-1'-[6-azanyl-5-(2-azanyl-3-chloranyl-pyridin-4-yl)sulfanyl-pyrazin-2-yl]spiro[5,7-dihydrocyclopenta[b]pyridine-6,4'-piperidine]-5-amine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Ma, C, Gao, P, Kang, D, Han, H, Sun, X, Zhang, W, Qian, D, Wang, Y, Long, W. | | Deposit date: | 2023-10-27 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of JAB-3312, a Potent SHP2 Allosteric Inhibitor for Cancer Treatment.
J.Med.Chem., 67, 2024
|
|
6J36
 
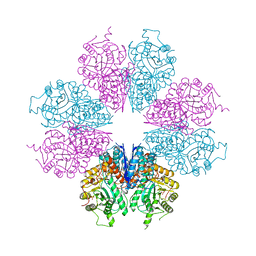 | | crystal structure of Mycoplasma hyopneumoniae Enolase | | Descriptor: | Enolase, GLYCEROL, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Xie, X, Feng, Z, Wang, W, Ran, T, Zhang, W, Xiang, Q, Shao, G. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Featured Species-Specific Loops Are Found in the Crystal Structure ofMhpEno, a Cell Surface Adhesin FromMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 9, 2019
|
|
8K20
 
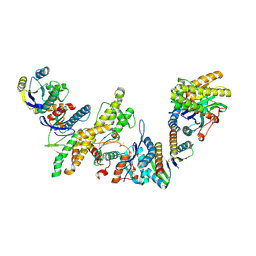 | | Cryo-EM structure of KEOPS complex from Arabidopsis thaliana | | Descriptor: | At4g34412, At5g53043, FE (III) ION, ... | | Authors: | Zheng, X.X, Zhu, L, Duan, L, Zhang, W.H. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis of A. thaliana KEOPS complex in biosynthesizing tRNA t6A.
Nucleic Acids Res., 52, 2024
|
|
2MA1
 
 | | Solution structure of HRDC1 domain of RecQ helicase from Deinococcus radiodurans | | Descriptor: | DNA helicase RecQ | | Authors: | Liu, S, Zhang, W, Gao, Z, Ming, Q, Hou, H, Lan, W, Wu, H, Cao, C, Dong, Y. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal-most HRDC1 domain of RecQ helicase from Deinococcus radiodurans
To be Published
|
|
5VKQ
 
 | | Structure of a mechanotransduction ion channel Drosophila NOMPC in nanodisc | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, No mechanoreceptor potential C isoform L | | Authors: | Jin, P, Bulkley, D, Guo, Y, Zhang, W, Guo, Z, Huynh, W, Wu, S, Meltzer, S, Chen, T, Jan, L.Y, Jan, Y.-N, Cheng, Y. | | Deposit date: | 2017-04-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Electron cryo-microscopy structure of the mechanotransduction channel NOMPC.
Nature, 547, 2017
|
|
5HCK
 
 | | HUMAN HCK SH3 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEMATOPOIETIC CELL KINASE | | Authors: | Horita, D.A, Baldisseri, D.M, Zhang, W, Altieri, A.S, Smithgall, T.E, Gmeiner, W.H, Byrd, R.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human Hck SH3 domain and identification of its ligand binding site.
J.Mol.Biol., 278, 1998
|
|
7U0N
 
 | | Crystal structure of chimeric omicron RBD (strain BA.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Q, Shi, K, Ye, G, Zhang, W, Aihara, H, Li, F. | | Deposit date: | 2022-02-18 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for Human Receptor Recognition by SARS-CoV-2 Omicron Variant BA.1.
J.Virol., 96, 2022
|
|
5VCI
 
 | | RNA hairpin structure containing tetraloop/receptor motif, complexed with 2-MeImpG analogue | | Descriptor: | 5'-O-[(S)-hydroxy(4-methyl-1H-imidazol-5-yl)phosphoryl]guanosine, RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*CP*CP*CP*GP*AP*AP*AP*GP*G)-3'), RNA (5'-R(P*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2017-03-31 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Rationale for the Enhanced Catalysis of Nonenzymatic RNA Primer Extension by a Downstream Oligonucleotide.
J. Am. Chem. Soc., 140, 2018
|
|
7N7X
 
 | | Crystal structure of BCX7353(ORLADEYO) in complex with human plasma kallikrein serine protease domain at 2.1 angstrom resolution | | Descriptor: | Orladeyo, PHOSPHATE ION, Plasma kallikrein light chain | | Authors: | Krishnan, R, Yarlagadda, B.S, Kotian, P, Polach, K.J, Zhang, W. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Berotralstat (BCX7353): Structure-Guided Design of a Potent, Selective, and Oral Plasma Kallikrein Inhibitor to Prevent Attacks of Hereditary Angioedema (HAE).
J.Med.Chem., 64, 2021
|
|
8XIF
 
 | | The crystal structure of the AEP domain of VACV D5 | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, Uncoating factor OPG117 | | Authors: | Gan, J, Zhang, W. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8XIG
 
 | | The crystal structure of the AEP domain of MPXV E5 | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, Uncoating factor OPG117 | | Authors: | Gan, J, Zhang, W. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
2JZ1
 
 | | DSX_long | | Descriptor: | Protein doublesex | | Authors: | Yang, Y, Zhang, W, Bayrer, J.R, Weiss, M.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Doublesex and the Regulation of Sexual Dimorphism in Drosophila melanogaster: STRUCTURE, FUNCTION, AND MUTAGENESIS OF A FEMALE-SPECIFIC DOMAIN.
J.Biol.Chem., 283, 2008
|
|
8YB7
 
 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C3 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
8YAX
 
 | | SARS-CoV-2 DMV nsp3-4 pore complex (full-pore) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-10 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
8YB5
 
 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C6 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
7M3Q
 
 | | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, ... | | Authors: | Chowdhury, A, Singer, A.U, Ogunjimi, A.A, Teyra, J, Zhang, W, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-03-18 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV)
To be published
|
|
2JZ0
 
 | | DSX_short | | Descriptor: | Protein doublesex | | Authors: | Yang, Y, Zhang, W, Bayrer, J.R, Weiss, M.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Doublesex and the Regulation of Sexual Dimorphism in Drosophila melanogaster: STRUCTURE, FUNCTION, AND MUTAGENESIS OF A FEMALE-SPECIFIC DOMAIN.
J.Biol.Chem., 283, 2008
|
|
3CFV
 
 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | Descriptor: | ARSENIC, Histone H4 peptide, Histone-binding protein RBBP7 | | Authors: | Pei, X.-Y, Murzina, N.V, Zhang, W, McLaughlin, S, Verreault, A, Luisi, B.F, Laue, E.D. | | Deposit date: | 2008-03-04 | | Release date: | 2008-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
2L85
 
 | | Solution NMR structures of CBP bromodomain with small molecule of HBS | | Descriptor: | 4-[(E)-(4-hydroxyphenyl)diazenyl]benzenesulfonic acid, CREB-binding protein | | Authors: | Borah, J.C, Mujtaba, S, Karakikes, I, Zeng, L, Muller, M, Patel, J, Moshkina, N, Morohashi, K, Zhang, W, Gerona-Navarro, G, Hajjar, R.J, Zhou, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Binding to the Coactivator CREB-Binding Protein Blocks Apoptosis in Cardiomyocytes.
Chem.Biol., 18, 2011
|
|
