5YK0
 
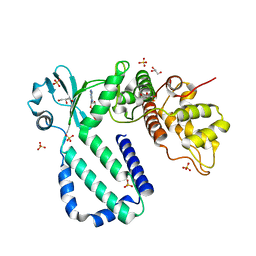 | | The complex structure of Rv3197-ADP from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Probable conserved ATP-binding protein ABC transporter, ... | | Authors: | Rao, Z.H, Zhang, Q.Q. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
5YK2
 
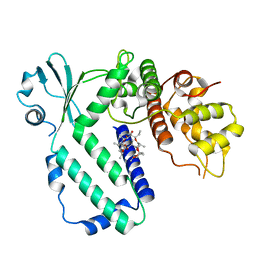 | |
3ME6
 
 | | Crystal structure of cytochrome 2B4 in complex with the anti-platelet drug clopidogrel | | Descriptor: | Clopidogrel, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Roberts, A.G, Maekawa, K, Talakad, J.C, Hong, W.X, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2010-03-31 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel.
Biochemistry, 49, 2010
|
|
3JBM
 
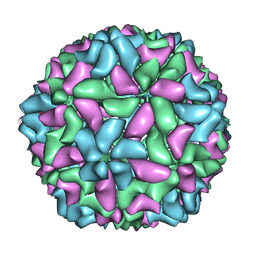 | | Electron cryo-microscopy of a virus-like particle of orange-spotted grouper nervous necrosis virus | | Descriptor: | virus-like particle of orange-spotted grouper nervous necrosis virus | | Authors: | Xie, J, Li, K, Gao, Y, Huang, R, Lai, Y, Shi, Y, Yang, S, Zhu, G, Zhang, Q, He, J. | | Deposit date: | 2015-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle
Vet. Res., 47, 2016
|
|
3K9Y
 
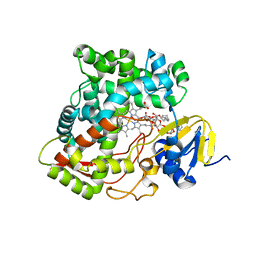 | | Crystal structure of rat mitochondrial P450 24A1 S57D in complex with CYMAL-5 | | Descriptor: | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, ... | | Authors: | Annalora, A.J, Goodin, D.B, Hong, W, Zhang, Q, Johnson, E.F, Stout, C.D. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism.
J.Mol.Biol., 396, 2010
|
|
3JA9
 
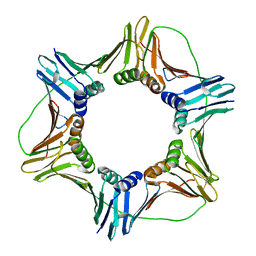 | |
3JAA
 
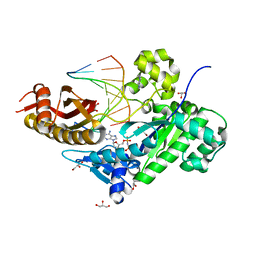 | | HUMAN DNA POLYMERASE ETA in COMPLEX WITH NORMAL DNA AND INCO NUCLEOTIDE (NRM) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*T*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3, DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Lau, W.C.Y, Li, Y, Zhang, Q, Huen, M.S.Y. | | Deposit date: | 2015-05-19 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Molecular architecture of the Ub-PCNA/Pol eta complex bound to DNA
Sci Rep, 5, 2015
|
|
3IXV
 
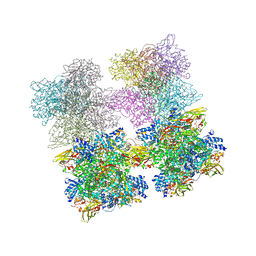 | | Scorpion Hemocyanin resting state pseudo atomic model built based on cryo-EM density map | | Descriptor: | Hemocyanin AA6 chain | | Authors: | Cong, Y, Zhang, Q, Woolford, D, Schweikardt, T, Khant, H, Ludtke, S, Chiu, W, Decker, H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural Mechanism of SDS-Induced Enzyme Activity of Scorpion Hemocyanin Revealed by Electron Cryomicroscopy.
Structure, 17, 2009
|
|
3IXW
 
 | | Scorpion Hemocyanin activated state pseudo atomic model built based on cryo-EM density map | | Descriptor: | Hemocyanin AA6 chain | | Authors: | Cong, Y, Zhang, Q, Woolford, D, Schweikardt, T, Khant, H, Ludtke, S, Chiu, W, Decker, H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural Mechanism of SDS-Induced Enzyme Activity of Scorpion Hemocyanin Revealed by Electron Cryomicroscopy.
Structure, 17, 2009
|
|
3J40
 
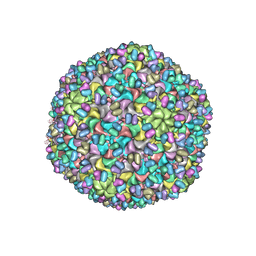 | | Validated Near-Atomic Resolution Structure of Bacteriophage Epsilon15 Derived from Cryo-EM and Modeling | | Descriptor: | gp10, gp7 | | Authors: | Baker, M.L, Hryc, C.F, Zhang, Q, Wu, W, Jakana, J, Haase-Pettingell, C, Afonine, P.V, Adams, P.D, King, J.A, Jiang, W, Chiu, W. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5XPV
 
 | | Structure of the V domain of amphioxus IgVJ-C2 | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
4HYX
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-4 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, DECANE, GLYCEROL, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-14 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3KW4
 
 | | Crystal structure of cytochrome 2B4 in complex with the anti-platelet drug ticlopidine | | Descriptor: | 2-{[(3alpha,5alpha,7alpha,8alpha,10alpha,12alpha,17alpha)-3,12-bis{2-[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]ethoxy}cholan-7-yl]oxy}ethyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Maekawa, K, Roberts, A.G, Hong, W.-X, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2009-11-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel.
Biochemistry, 49, 2010
|
|
4TVR
 
 | | Tandem Tudor and PHD domains of UHRF2 | | Descriptor: | E3 ubiquitin-protein ligase UHRF2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Walker, J.R, Dong, A, Zhang, Q, Ong, M, Duan, S, Li, Y, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of the Tandem Tudor and PHD domains of UHRF2
To be published
|
|
1B8Q
 
 | | SOLUTION STRUCTURE OF THE EXTENDED NEURONAL NITRIC OXIDE SYNTHASE PDZ DOMAIN COMPLEXED WITH AN ASSOCIATED PEPTIDE | | Descriptor: | PROTEIN (HEPTAPEPTIDE), PROTEIN (NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Tochio, H, Zhang, Q, Mandal, P, Li, M, Zhang, M. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide.
Nat.Struct.Biol., 6, 1999
|
|
3LJW
 
 | | Crystal Structure of the Second Bromodomain of Human Polybromo | | Descriptor: | ACETATE ION, Protein polybromo-1, SODIUM ION | | Authors: | Charlop-Powers, Z, Zhou, M.M, Zeng, L, Zhang, Q. | | Deposit date: | 2010-01-26 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2.
Cell Res., 20, 2010
|
|
3QPS
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3QQA
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.-C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
6BL6
 
 | | Crystallization of lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Stanfield, R.L, Zhang, Q, Wilson, I.A, Lee, S.C. | | Deposit date: | 2017-11-09 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
4HWL
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-7 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, GLYCEROL, HEPTANE, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1DV5
 
 | | TERTIARY STRUCTURE OF APO-D-ALANYL CARRIER PROTEIN | | Descriptor: | APO-D-ALANYL CARRIER PROTEIN | | Authors: | Volkman, B.F, Zhang, Q, Debabov, D.V, Rivera, E, Kresheck, G.C, Neuhaus, F.C. | | Deposit date: | 2000-01-19 | | Release date: | 2001-08-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Biosynthesis of D-alanyl-lipoteichoic acid: the tertiary structure of apo-D-alanyl carrier protein.
Biochemistry, 40, 2001
|
|
7ZKA
 
 | | ABCB1 V978C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Urbatsch, I, Zhang, Q, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK9
 
 | | ABCB1 L971C mutant (mABCB1) in the inward facing state | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | Authors: | Parey, K, Januliene, D, Gewering, T, Zhang, Q, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
