6UBZ
 
 | | Crystal structure of D678A GoxA bound to glycine at pH 5.5 | | Descriptor: | GLYCINE, MAGNESIUM ION, Uncharacterized protein GoxA | | Authors: | Yukl, E.T. | | Deposit date: | 2019-09-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Kinetic and structural evidence that Asp-678 plays multiple roles in catalysis by the quinoprotein glycine oxidase.
J.Biol.Chem., 294, 2019
|
|
6UC1
 
 | | Crystal structure of D678A GoxA soaked in glycine at pH 7.5 | | Descriptor: | GLYCINE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Yukl, E.T. | | Deposit date: | 2019-09-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Kinetic and structural evidence that Asp-678 plays multiple roles in catalysis by the quinoprotein glycine oxidase.
J.Biol.Chem., 294, 2019
|
|
6VMF
 
 | | Crystal structure of the Y766F mutant of GoxA soaked with glycine | | Descriptor: | Glycine oxidase, MAGNESIUM ION, SULFATE ION | | Authors: | Yukl, E.T. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Roles of active-site residues in catalysis, substrate binding, cooperativity, and the reaction mechanism of the quinoprotein glycine oxidase.
J.Biol.Chem., 295, 2020
|
|
6VMV
 
 | | Crystal structure of the H767A mutant of GoxA soaked with glycine | | Descriptor: | Glycine oxidase, MAGNESIUM ION, SULFATE ION | | Authors: | Yukl, E.T. | | Deposit date: | 2020-01-28 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Roles of active-site residues in catalysis, substrate binding, cooperativity, and the reaction mechanism of the quinoprotein glycine oxidase.
J.Biol.Chem., 295, 2020
|
|
6VMW
 
 | | Crystal structure of the F316A mutant of GoxA soaked with glycine | | Descriptor: | Glycine oxidase, MAGNESIUM ION, SODIUM ION | | Authors: | Yukl, E.T. | | Deposit date: | 2020-01-28 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Roles of active-site residues in catalysis, substrate binding, cooperativity, and the reaction mechanism of the quinoprotein glycine oxidase.
J.Biol.Chem., 295, 2020
|
|
6VL7
 
 | | Crystal structure of the H583C mutant of GoxA soaked with glycine | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glycine oxidase, MAGNESIUM ION, ... | | Authors: | Yukl, E.T. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Roles of active-site residues in catalysis, substrate binding, cooperativity, and the reaction mechanism of the quinoprotein glycine oxidase.
J.Biol.Chem., 295, 2020
|
|
5F81
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2015-12-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
8F1B
 
 | |
8EZW
 
 | |
5W57
 
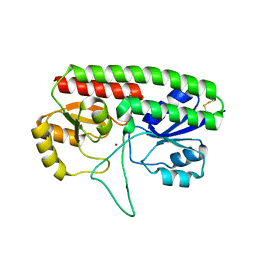 | | Structure of Holo AztC | | Descriptor: | Periplasmic solute binding protein, ZINC ION | | Authors: | Avalos, D, Yukl, E.T. | | Deposit date: | 2017-06-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of zinc binding to the solute-binding protein AztC and transfer from the metallochaperone AztD.
J. Biol. Chem., 292, 2017
|
|
5W56
 
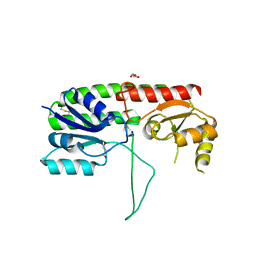 | | Structure of Apo AztC | | Descriptor: | GLYCEROL, Periplasmic solute binding protein, SODIUM ION | | Authors: | Avalos, D, Yukl, E.T. | | Deposit date: | 2017-06-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanisms of zinc binding to the solute-binding protein AztC and transfer from the metallochaperone AztD.
J. Biol. Chem., 292, 2017
|
|
5HL4
 
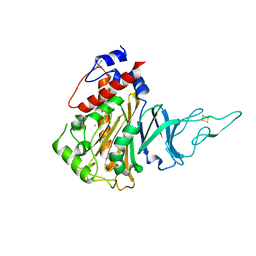 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | COBALT HEXAMMINE(III), FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
5HQD
 
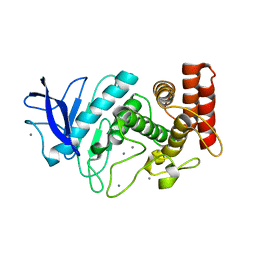 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Roesser, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-21 | | Release date: | 2016-02-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
4KFD
 
 | | Crystal structure of Hansenula polymorpha copper amine oxidase-1 reduced by methylamine at pH 6.0 | | Descriptor: | COPPER (II) ION, GLYCEROL, HYDROGEN PEROXIDE, ... | | Authors: | Johnson, B.J, Yukl, E.T, Klema, V.J, Wilmot, C.M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural evidence for the semiquinone in a copper amine oxidase from Hansenula polymorpha: implications for the catalytic mechanism
J.Biol.Chem., 2013
|
|
4KFF
 
 | | Crystal structure of Hansenula polymorpha copper amine oxidase-1 reduced by methylamine at pH 8.5 | | Descriptor: | COPPER (II) ION, GLYCEROL, Peroxisomal primary amine oxidase | | Authors: | Johnson, B.J, Yukl, E.T, Klema, V.J, Wilmot, C.M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural evidence for the semiquinone in a copper amine oxidase from Hansenula polymorpha: implications for the catalytic mechanism
J.Biol.Chem., 2013
|
|
4KFE
 
 | | Crystal structure of Hansenula polymorpha copper amine oxidase-1 reduced by methylamine at pH 7.0 | | Descriptor: | COPPER (II) ION, FORMYL GROUP, GLYCEROL, ... | | Authors: | Johnson, B.J, Yukl, E.T, Klema, V.J, Wilmot, C.M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for the semiquinone in a copper amine oxidase from Hansenula polymorpha: implications for the catalytic mechanism
J.Biol.Chem., 2013
|
|
