6PTJ
 
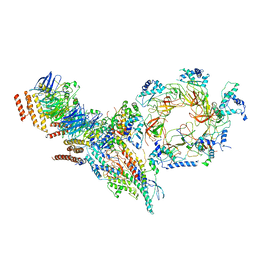 | | Structure of Ctf4 trimer in complex with one CMG helicase | | Descriptor: | Cell division control protein 45, DNA polymerase alpha-binding protein, DNA replication complex GINS protein PSF1, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PTN
 
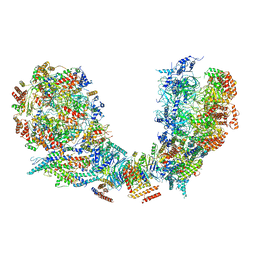 | | Structure of Ctf4 trimer in complex with two CMG helicases | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PTO
 
 | | Structure of Ctf4 trimer in complex with three CMG helicases | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
9D9V
 
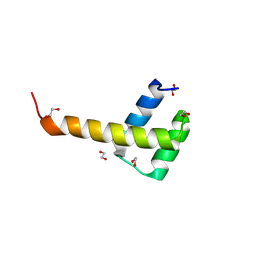 | | X-Ray structure of ALX4 homeodomain | | Descriptor: | 1,2-ETHANEDIOL, Homeobox protein aristaless-like 4 | | Authors: | Yuan, Z, Kovall, R.A. | | Deposit date: | 2024-08-21 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and biochemical characterization of the ALX4 dimer reveals novel insights into how disease alleles impact ALX4 function
To Be Published
|
|
9D9R
 
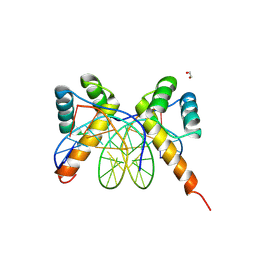 | | X-ray structure of ALX4 homeodomain dimer bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*AP*TP*TP*CP*AP*AP*TP*TP*AP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*TP*AP*AP*TP*TP*GP*AP*AP*TP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Yuan, Z, Kovall, R.A. | | Deposit date: | 2024-08-21 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | Structural and biochemical characterization of the ALX4 dimer reveals novel insights into how disease alleles impact ALX4 function
To Be Published
|
|
6U0M
 
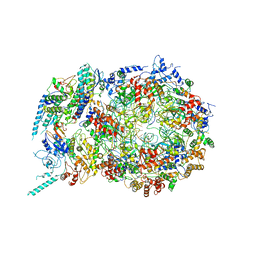 | | Structure of the S. cerevisiae replicative helicase CMG in complex with a forked DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (15-MER), ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Zhang, D, O'Donnell, M, Li, H. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA unwinding mechanism of a eukaryotic replicative CMG helicase.
Nat Commun, 11, 2020
|
|
9IQX
 
 | |
9IQY
 
 | |
8FOJ
 
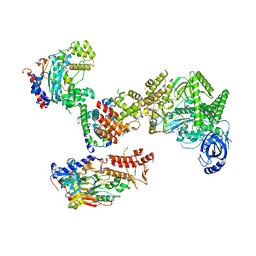 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the post RNA handoff state | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOC
 
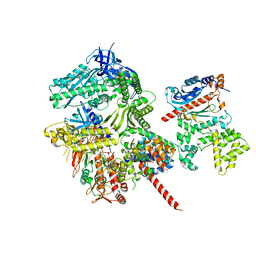 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase in Apo state conformation I | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOH
 
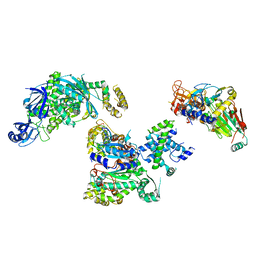 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the RNA synthesis state | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOE
 
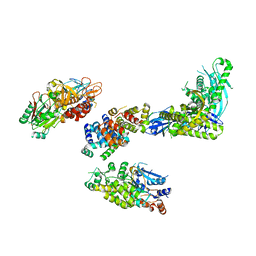 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex bound to a template DNA | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOK
 
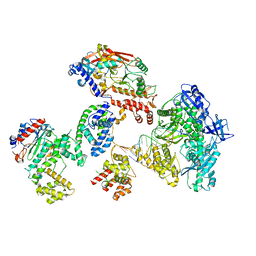 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the DNA elongation state | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
8FOD
 
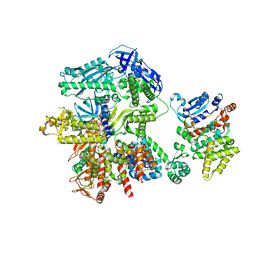 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in Apo state conformation II | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
6WJV
 
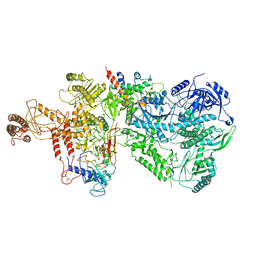 | | Structure of the Saccharomyces cerevisiae polymerase epsilon holoenzyme | | Descriptor: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit B, DNA polymerase epsilon subunit C, ... | | Authors: | Yuan, Z, Georgescu, R, Schauer, G.D, O'Donnell, M, Li, H. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the polymerase epsilon holoenzyme and atomic model of the leading strand replisome.
Nat Commun, 11, 2020
|
|
6WGG
 
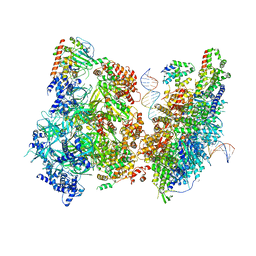 | | Atomic model of pre-insertion mutant OCCM-DNA complex(ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (41-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGC
 
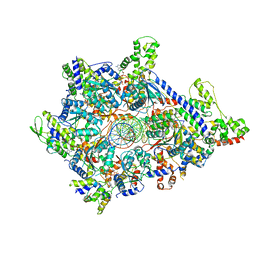 | | Atomic model of semi-attached mutant OCCM-DNA complex (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, DNA (41-MER), DNA replication licensing factor MCM3, ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGF
 
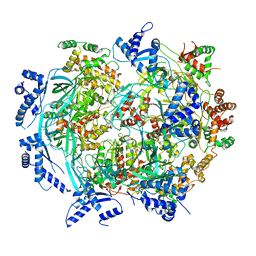 | | Atomic model of mutant Mcm2-7 hexamer with Mcm6 WHD truncation | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGI
 
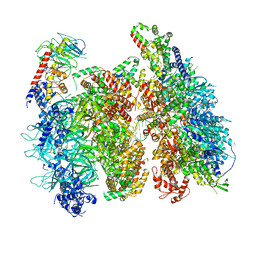 | | Atomic model of the mutant OCCM (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) loaded on DNA at 10.5 A resolution | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (34-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
9B8R
 
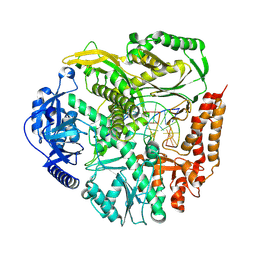 | | Cryo-EM structure of S. cerevisiae PolE-core-DNA | | Descriptor: | DNA polymerase epsilon catalytic subunit, IRON/SULFUR CLUSTER, Primer DNA, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2024-03-31 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis.
Science, 385, 2024
|
|
6A06
 
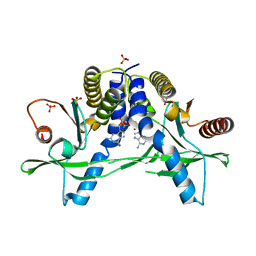 | | Structure of pSTING complex | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A04
 
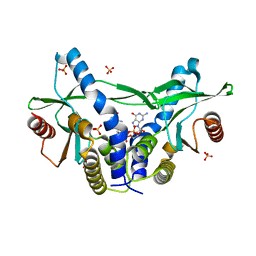 | | Structure of pSTING complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A05
 
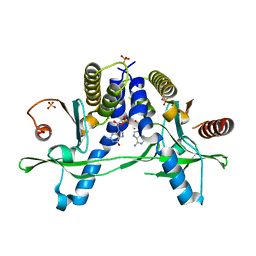 | | Structure of pSTING complex | | Descriptor: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A03
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
8TWB
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis.
Science, 385, 2024
|
|
