8CR9
 
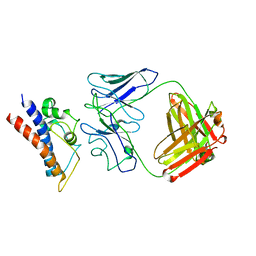 | | Cryo-EM structure of PcrV/Fab(30-B8) | | Descriptor: | Heavy chain, Maltose/maltodextrin-binding periplasmic protein,Type III secretion protein PcrV, light chain | | Authors: | Yuan, B, Simonis, A, Marlovits, T.C. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Discovery of highly neutralizing human antibodies targeting Pseudomonas aeruginosa.
Cell, 186, 2023
|
|
8CRB
 
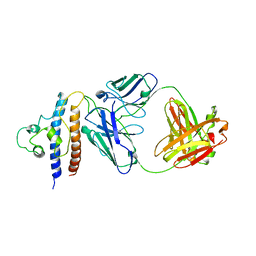 | | Cryo-EM structure of PcrV/Fab(11-E5) | | Descriptor: | Heavy chain, Light chain, Maltose/maltodextrin-binding periplasmic protein,Type III secretion protein PcrV | | Authors: | Yuan, B, Simonis, A, Marlovits, T.C. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Discovery of highly neutralizing human antibodies targeting Pseudomonas aeruginosa.
Cell, 186, 2023
|
|
8C4C
 
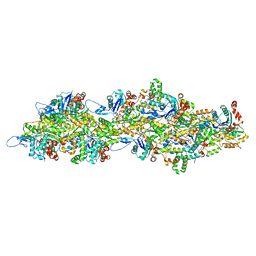 | | F-actin decorated by SipA497-669 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Yuan, B, Wald, J, Marlovits, T.C. | | Deposit date: | 2023-01-03 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for subversion of host cell actin cytoskeleton during Salmonella infection.
Sci Adv, 9, 2023
|
|
8C4E
 
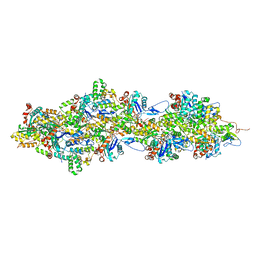 | | F-actin decorated by SipA426-685 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Yuan, B, Wald, J, Marlovits, T.C. | | Deposit date: | 2023-01-03 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for subversion of host cell actin cytoskeleton during Salmonella infection.
Sci Adv, 9, 2023
|
|
8ADG
 
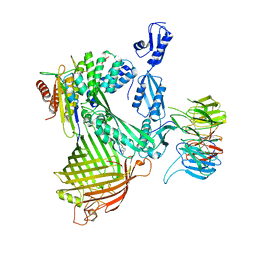 | | Cryo-EM structure of Darobactin 22 bound BAM complex | | Descriptor: | Darobactin 22, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Yuan, B, Marlovits, T.C. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ADI
 
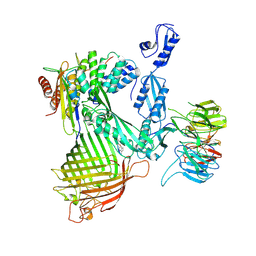 | | Cryo-EM structure of Darobactin 9 bound BAM complex | | Descriptor: | Darobactin 9, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Yuan, B, Marlovits, T.C. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7OSL
 
 | |
7YER
 
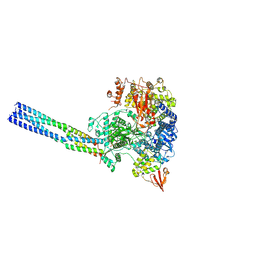 | | The structure of EBOV L-VP35 complex | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
7YET
 
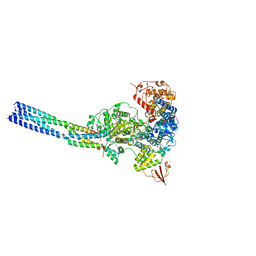 | | The structure of EBOV L-VP35 in complex with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Polymerase cofactor VP35, RNA-directed RNA polymerase L | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
7YES
 
 | |
