2DHR
 
 | | Whole cytosolic region of ATP-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
2DI4
 
 | | Crystal structure of the FtsH protease domain | | Descriptor: | Cell division protein ftsH homolog, MERCURY (II) ION | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-28 | | Release date: | 2006-06-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
2ENM
 
 | | Solution structure of the SH3 domain from mouse sorting nexin-9 | | Descriptor: | Sorting nexin-9 | | Authors: | Wakabayashi, M, Kurosaki, C, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from mouse sorting nexin-9
To be Published
|
|
3APD
 
 | | Crystal structure of human PI3K-gamma in complex with CH5108134 | | Descriptor: | 5-(2-Morpholin-4-yl-7-pyridin-3-yl-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl)-pyrimidin-2-ylamine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Nakamura, M, Fukami, T.A, Miyazaki, T, Yoshida, M. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3APC
 
 | | Crystal structure of human PI3K-gamma in complex with CH5132799 | | Descriptor: | 5-(7-Methanesulfonyl-2-morpholin-4-yl-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl)-pyrimidin-2-ylamine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Nakamura, M, Fukami, T.A, Miyazaki, T, Yoshida, M. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.544 Å) | | Cite: | Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3APF
 
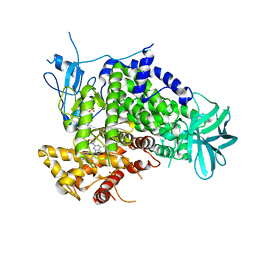 | | Crystal structure of human PI3K-gamma in complex with CH5039699 | | Descriptor: | 3-[7-(1H-benzimidazol-5-yl)-2-(morpholin-4-yl)-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl]phenol, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Nakamura, M, Fukami, T.A, Miyazaki, T, Yoshida, M. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2EDF
 
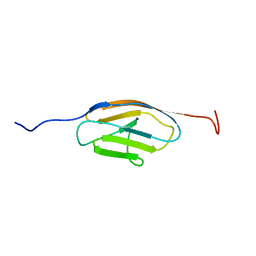 | |
2ENS
 
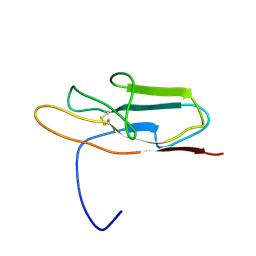 | | Solution structure of the third ig-like domain from human Advanced glycosylation end product-specific receptor | | Descriptor: | Advanced glycosylation end product-specific receptor | | Authors: | Wakabayashi, M, Kurosaki, C, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2008-04-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third ig-like domain from human Advanced glycosylation end product-specific receptor
To be Published
|
|
1IY2
 
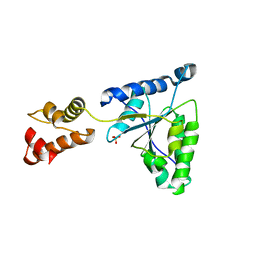 | | Crystal structure of the FtsH ATPase domain from Thermus thermophilus | | Descriptor: | ATP-dependent metalloprotease FtsH, SULFATE ION | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1J3C
 
 | |
1J3X
 
 | |
1IY0
 
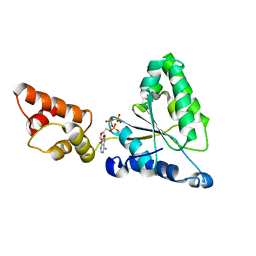 | | Crystal structure of the FtsH ATPase domain with AMP-PNP from Thermus thermophilus | | Descriptor: | ATP-dependent metalloprotease FtsH, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1L5H
 
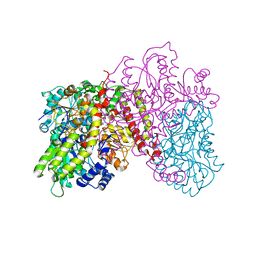 | | FeMo-cofactor Deficient Nitrogenase MoFe Protein | | Descriptor: | CALCIUM ION, FE(8)-S(7) CLUSTER, nitrogenase molybdenum-iron protein alpha chain, ... | | Authors: | Schmid, B, Ribbe, M.W, Einsle, O, Yoshida, M, Thomas, L.M, Dean, D.R, Rees, D.C, Burgess, B.K. | | Deposit date: | 2002-03-06 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a cofactor-deficient nitrogenase MoFe protein.
Science, 296, 2002
|
|
1IXZ
 
 | | Crystal structure of the FtsH ATPase domain from Thermus thermophilus | | Descriptor: | ATP-dependent metalloprotease FtsH, MERCURY (II) ION, SULFATE ION | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1M1Y
 
 | | Chemical Crosslink of Nitrogenase MoFe Protein and Fe Protein | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Schmid, B, Einsle, O, Chiu, H.J, Willing, A, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-20 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical and Structural Characterization of the Crosslinked Complex of Nitrogenase: Comparison to the ADP-AlF4- Stabilized Structure
Biochemistry, 41, 2002
|
|
1IY1
 
 | | Crystal structure of the FtsH ATPase domain with ADP from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent metalloprotease FtsH | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IYG
 
 | | Solution structure of RSGI RUH-001, a Fis1p-like and CGI-135 homologous domain from a mouse cDNA | | Descriptor: | Hypothetical protein (2010003O14) | | Authors: | Ohashi, W, Hirota, H, Yamazaki, T, Koshiba, S, Hamada, T, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-14 | | Release date: | 2003-02-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-001, a Fis1p-like and CGI-135 homologous domain from a mouse cDNA
To be Published
|
|
1J3D
 
 | |
1M34
 
 | | Nitrogenase Complex From Azotobacter Vinelandii Stabilized By ADP-Tetrafluoroaluminate | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Schmid, B, Einsle, O, Chiu, H.-J, Willing, A, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-27 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Characterization of the Crosslinked Complex of Nitrogenase: Comparison to the ADP-AlF4 Stabilized structure
Biochemistry, 41, 2002
|
|
1M1N
 
 | | Nitrogenase MoFe protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(7)-MO-S(9)-N CLUSTER, ... | | Authors: | Einsle, O, Tezcan, F.A, Andrade, S.L.A, Schmid, B, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-19 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Nitrogenase MoFe-protein at 1.16 A resolution: a central ligand in the FeMo-cofactor.
Science, 297, 2002
|
|
3W5S
 
 | | Crystal Structure of Maleylacetate Reductase from Rhizobium sp. strain MTP-10005 | | Descriptor: | BENZAMIDINE, GLYCEROL, Maleylacetate reductase, ... | | Authors: | Fujii, T, Ogawa, A, Goda, Y, Yamauchi, T, Yoshida, M, Oikawa, T, Hata, Y. | | Deposit date: | 2013-02-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of Maleylacetate Reductase from Rhizobium sp. strain MTP-10005
To be Published
|
|
1VB7
 
 | |
1VEG
 
 | | Solution Structure of RSGI RUH-012, a UBA Domain from Mouse cDNA | | Descriptor: | NEDD8 ultimate buster-1 | | Authors: | Abe, T, Hirota, H, Izumi, K, Yoshida, M, Yamazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-31 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-012, a UBA Domain from Mouse cDNA
To be Published
|
|
2D9C
 
 | |
2D9E
 
 | | Solution structure of the Bromodomain of Peregrin | | Descriptor: | Peregrin | | Authors: | Hatta, R, Hayashi, F, Izumi, K, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-09 | | Release date: | 2006-06-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Bromodomain of Peregrin
To be published
|
|
