4EN8
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN9
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactosamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN7
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactosamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
2EIS
 
 | | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8 | | Descriptor: | COENZYME A, Hypothetical protein TTHB207 | | Authors: | Kamitori, S, Yoshida, H, Satoh, S, Iino, H, Ebihara, A, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8
To be Published
|
|
1RCK
 
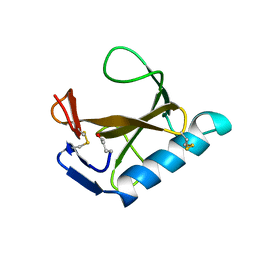 | |
1RCL
 
 | |
5B23
 
 | |
2D0F
 
 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N complexed with P2, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2D0G
 
 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P5, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2D2O
 
 | | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft | | Descriptor: | CALCIUM ION, Neopullulanase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ohtaki, A, Mizuno, M, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-09-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft
Carbohydr.Res., 341, 2006
|
|
2D0H
 
 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P2, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2ECE
 
 | |
5ZQT
 
 | | Crystal structure of Oryza sativa hexokinase 6 | | Descriptor: | Hexokinase-6, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsudaira, K, Mochizuki, S, Yoshida, H, Kamitori, S, Akimitsu, K. | | Deposit date: | 2018-04-20 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of Oryza sativa hexokinase 6
To Be Published
|
|
2Z8G
 
 | | Aspergillus niger ATCC9642 isopullulanase complexed with isopanose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Mizuno, M, Koide, A, Yamamura, A, Akeboshi, H, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2007-09-05 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
3VUS
 
 | | Escherichia coli PgaB N-terminal domain | | Descriptor: | ACETATE ION, MERCURY (II) ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase, ... | | Authors: | Nishiyama, T, Noguchi, H, Yoshida, H, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-07-05 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: a circularly permuted member of the carbohydrate esterase 4 family
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2ZYK
 
 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with gamma-cyclodextrin | | Descriptor: | Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Tonozuka, T, Sogawa, A, Yamada, M, Matsumoto, N, Yoshida, H, Kamitori, S, Ichikawa, K, Mizuno, M, Nishikawa, A, Sakano, Y. | | Deposit date: | 2009-01-26 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cyclodextrin recognition by Thermoactinomyces vulgaris cyclo/maltodextrin-binding protein
Febs J., 274, 2007
|
|
3WUC
 
 | | X-ray crystal structure of Xenopus laevis galectin-Va | | Descriptor: | Galectin, MALONIC ACID, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nonaka, Y, Yoshida, H, Kamitori, S, Nakamura, T. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a Xenopus laevis skin proto-type galectin, close to but distinct from galectin-1.
Glycobiology, 25, 2015
|
|
3WUD
 
 | | X-ray crystal structure of Xenopus laevis galectin-Ib | | Descriptor: | Galectin, SULFATE ION, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nonaka, Y, Yoshida, H, Kamitori, S, Nakamura, T. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of a Xenopus laevis skin proto-type galectin, close to but distinct from galectin-1.
Glycobiology, 25, 2015
|
|
2ZKN
 
 | |
3VKN
 
 | | Galectin-8 N-terminal domain in free form | | Descriptor: | CHLORIDE ION, Galectin-8 | | Authors: | Kamitori, S, Yoshida, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
3VKL
 
 | |
3VKO
 
 | | Galectin-8 N-terminal domain in complex with sialyllactosamine | | Descriptor: | CHLORIDE ION, Galectin-8, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kamitori, S, Yoshida, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
3WW3
 
 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with no ligand | | Descriptor: | L-ribose isomerase, MANGANESE (II) ION | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3W7S
 
 | | Escherichia coli K12 YgjK complexed with glucose | | Descriptor: | CALCIUM ION, Uncharacterized protein YgjK, alpha-D-glucopyranose | | Authors: | Miyazaki, T, Kurakata, Y, Uechi, A, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-06 | | Release date: | 2013-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63.
J.Mol.Biol., 381, 2008
|
|
3WW1
 
 | | X-ray structure of Cellulomonas parahominis L-ribose isomerase with L-ribose | | Descriptor: | L-ribose, L-ribose isomerase, MANGANESE (II) ION, ... | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
