5WQW
 
 | | X-ray structure of catalytic domain of autolysin from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, N-acetylglucosaminidase | | Authors: | Tamai, E, Sekiya, H, Goda, E, Makihata, N, Maki, J, Yoshida, H, Kamitori, S. | | Deposit date: | 2016-11-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and biochemical characterization of the Clostridium perfringens autolysin catalytic domain
FEBS Lett., 591, 2017
|
|
5X2J
 
 | | Crystal structure of a recombinant hybrid manganese superoxide dismutase from Staphylococcus equorum and Staphylococcus saprophyticus | | Descriptor: | MANGANESE (II) ION, manganese superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Arumsari, S, Kamitori, S, Ismaya, W.T. | | Deposit date: | 2017-01-31 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of manganese superoxide dismutase from the genus Staphylococcus
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6M30
 
 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase N73F | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Razani, M.D, Meidianto, V.F, Hartanto, A, Artarini, A, Ismaya, W.T. | | Deposit date: | 2020-03-02 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Unprecedented Role of The N73-F124 Pair in The Staphylococcus equorum MnSOD Activity.
Curr Enzym Inhib, 2021
|
|
3CW4
 
 | | Large c-terminal domain of influenza a virus RNA-dependent polymerase PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Kuzuhara, T, Kise, D, Yoshida, H, Horita, T, Murasaki, Y, Utsunomiya, H, Fujiki, H, Tsuge, H. | | Deposit date: | 2008-04-21 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the influenza A virus RNA polymerase PB2 RNA-binding domain containing the pathogenicity-determinant lysine 627 residue
J.Biol.Chem., 284, 2009
|
|
3D3I
 
 | | Crystal structural of Escherichia coli K12 YgjK, a glucosidase belonging to glycoside hydrolase family 63 | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein ygjK | | Authors: | Kurakata, Y, Uechi, A, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2008-05-12 | | Release date: | 2008-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63.
J.Mol.Biol., 381, 2008
|
|
7W6W
 
 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase L169W | | Descriptor: | AZIDE ION, MANGANESE (II) ION, Superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Artarini, A.A, Ismaya, W.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Introducing Intermolecular Interaction to Strengthen the Stability of MnSOD Dimer.
Appl.Biochem.Biotechnol., 195, 2023
|
|
7WNK
 
 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase K38R and A121E | | Descriptor: | AZIDE ION, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Retnoningrum, D.S, Yoshida, H, Artarini, A.A, Ismaya, W.T. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Introducing Intermolecular Interaction to Strengthen the Stability of MnSOD Dimer.
Appl.Biochem.Biotechnol., 195, 2023
|
|
7WNL
 
 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase K38R and A121Y | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, BROMIDE ION, ... | | Authors: | Retnoningrum, D.S, Yoshida, H, Artarini, A.A, Ismaya, W.T. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Introducing Intermolecular Interaction to Strengthen the Stability of MnSOD Dimer.
Appl.Biochem.Biotechnol., 195, 2023
|
|
4JY2
 
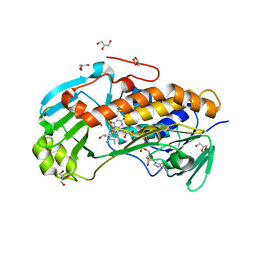 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, native and unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4KRU
 
 | |
4JY3
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-pyridoxic acid bound form | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxy-4-(hydroxymethyl)-6-methylpyridine-3-carboxylic acid, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-29 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4KRT
 
 | |
2X1D
 
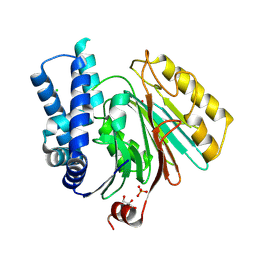 | | The crystal structure of mature acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, ACETATE ION, ACYL-COENZYME, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
2X1C
 
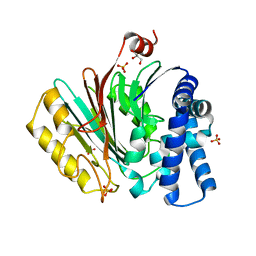 | | The crystal structure of precursor acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum | | Descriptor: | ACYL-COENZYME, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
5H34
 
 | | Crystal structure of the C-terminal domain of methionyl-tRNA synthetase (MetRS-C) in Nanoarchaeum equitans | | Descriptor: | Methionine-tRNA ligase | | Authors: | Suzuki, H, Kaneko, A, Yamamoto, T, Nambo, M, Umehara, T, Yoshida, H, Park, S.Y, Tamura, K. | | Deposit date: | 2016-10-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Binding Properties of Split tRNA to the C-terminal Domain of Methionyl-tRNA Synthetase of Nanoarchaeum equitans.
J. Mol. Evol., 84, 2017
|
|
7DDW
 
 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase S126C | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Razani, M.D, Meidianto, V.F, Hartanto, A, Artarini, A, Ismaya, W.T. | | Deposit date: | 2020-10-30 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The role of S126 in the Staphylococcus equorum MnSOD activity and stability.
J.Struct.Biol., 213, 2021
|
|
4H2P
 
 | | Tetrameric form of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4H2R
 
 | | Structure of MHPCO Y270F mutant, 5-hydroxynicotinic acid complex | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4GF7
 
 | | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO), unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Sawa, Y, Yagi, T. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4H2N
 
 | | Crystal structure of MHPCO, Y270F mutant | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4H2Q
 
 | | structure of MHPCO-5HN complex | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal structure of 2-Methyl-3-hydroxypyridiine-5-carboxylic acid oxygenase
To be Published
|
|
4EN8
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN9
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactosamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN7
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactosamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
2EIS
 
 | | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8 | | Descriptor: | COENZYME A, Hypothetical protein TTHB207 | | Authors: | Kamitori, S, Yoshida, H, Satoh, S, Iino, H, Ebihara, A, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8
To be Published
|
|
