3UMW
 
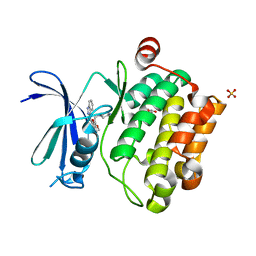 | | Crystal structure of Pim1 kinase in complex with inhibitor (Z)-2-[(1H-indazol-3-yl)methylene]-6-methoxy-7-(piperazin-1-ylmethyl)benzofuran-3(2H)-one | | Descriptor: | (2Z)-2-(1H-indazol-3-ylmethylidene)-6-methoxy-7-(piperazin-1-ylmethyl)-1-benzofuran-3(2H)-one, GLYCEROL, Proto-oncogene serine/threonine-protein kinase pim-1, ... | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Rational evolution of a novel type of potent and selective proviral integration site in Moloney murine leukemia virus kinase 1 (PIM1) inhibitor from a screening-hit compound.
J.Med.Chem., 55, 2012
|
|
3VYW
 
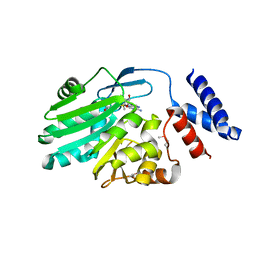 | | Crystal structure of MNMC2 from Aquifex Aeolicus | | Descriptor: | BENZAMIDINE, MNMC2, S-ADENOSYLMETHIONINE | | Authors: | Shibata, R, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization and structure of the Aquifex aeolicus protein DUF752: a bacterial tRNA-methyltransferase (MnmC2) functioning without the usually fused oxidase domain (MnmC1).
J.Biol.Chem., 287, 2012
|
|
3UMX
 
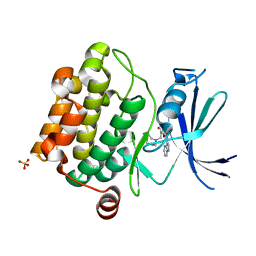 | | Crystal structure of Pim1 kinase in complex with inhibitor (Z)-2-[(1H-indol-3-yl)methylene]-7-(azepan-1-ylmethyl)-6-hydroxybenzofuran-3(2H)-one | | Descriptor: | (2Z)-7-(azepan-1-ylmethyl)-6-hydroxy-2-(1H-indol-3-ylmethylidene)-1-benzofuran-3(2H)-one, Proto-oncogene serine/threonine-protein kinase pim-1, SULFATE ION | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2011-11-15 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3WU6
 
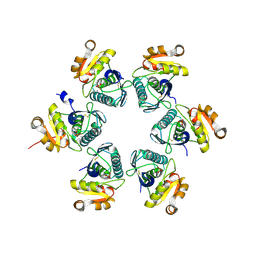 | | Oxidized E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
3VTA
 
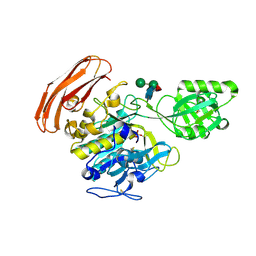 | | Crystal Structure of cucumisin, a subtilisin-like endoprotease from Cucumis melo L | | Descriptor: | Cucumisin, DIISOPROPYL PHOSPHONATE, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Murayama, K, Kato-Murayama, M, Hosaka, T, Sotokawauchi, A, Shirouzu, M, Arima, K, Yokoyama, S. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of cucumisin, a subtilisin-like endoprotease from Cucumis melo L
J.Mol.Biol., 423, 2012
|
|
3VHL
 
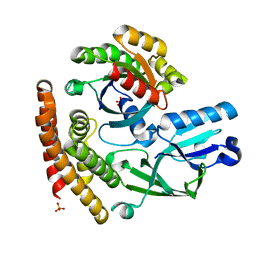 | | Crystal structure of the DHR-2 domain of DOCK8 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 8, PHOSPHATE ION | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Nishizak, T, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2011-08-26 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses.
Blood, 119, 2012
|
|
3VQX
 
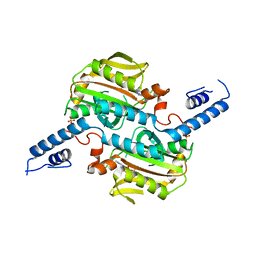 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in triclinic crystal form | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, Pyrrolysine--tRNA ligase, ... | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-02 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WPS
 
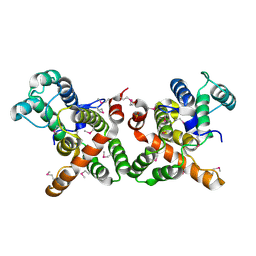 | | crystal structure of the GAP domain of MgcRacGAP(S387D) | | Descriptor: | Rac GTPase-activating protein 1, SULFATE ION | | Authors: | Murayama, K, Kato-murayama, M, Shirouzu, M, Kitamura, T, Yokoyama, S. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | crystal structure of the GAP domain of MgcRacGAP
To be Published
|
|
3WPQ
 
 | |
3WHE
 
 | | A new conserved neutralizing epitope at the globular head of hemagglutinin in H3N2 influenza viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fujii, Y, Sumida, T, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-08-25 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses.
J.Virol., 88, 2014
|
|
3WU5
 
 | | Reduced E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
3WJ9
 
 | |
3WU3
 
 | | Reduced-form structure of E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
3WU4
 
 | | Oxidized-form structure of E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
1UJ4
 
 | | Crystal structure of Thermus thermophilus ribose-5-phosphate isomerase | | Descriptor: | CHLORIDE ION, ribose 5-phosphate isomerase | | Authors: | Hamada, K, Ago, H, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxyanion hole-stabilized stereospecific isomerization in ribose-5-phosphate isomerase (Rpi)
J.Biol.Chem., 278, 2003
|
|
1UJ6
 
 | | Crystal structure of Thermus thermophilus ribose-5-phosphate isomerase complexed with arabinose-5-phosphate | | Descriptor: | ARABINOSE-5-PHOSPHATE, CHLORIDE ION, ribose 5-phosphate isomerase | | Authors: | Hamada, K, Ago, H, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Oxyanion hole-stabilized stereospecific isomerization in ribose-5-phosphate isomerase (Rpi)
J.Biol.Chem., 278, 2003
|
|
1UJ5
 
 | | Crystal structure of Thermus thermophilus ribose-5-phosphate isomerase complexed with ribose-5-phosphate | | Descriptor: | CHLORIDE ION, RIBULOSE-5-PHOSPHATE, ribose 5-phosphate isomerase | | Authors: | Hamada, K, Ago, H, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyanion hole-stabilized stereospecific isomerization in ribose-5-phosphate isomerase (Rpi)
J.Biol.Chem., 278, 2003
|
|
1UJP
 
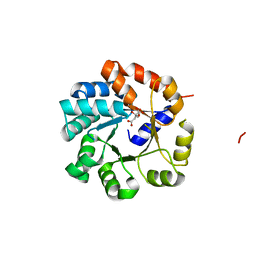 | | Crystal Structure of Tryptophan Synthase A-Subunit From Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, Tryptophan synthase alpha chain | | Authors: | Asada, Y, Yokoyama, S, Kuramitsu, S, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-08 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Stabilization mechanism of the tryptophan synthase alpha-subunit from Thermus thermophilus HB8: X-ray crystallographic analysis and calorimetry.
J.Biochem.(Tokyo), 138, 2005
|
|
2FLF
 
 | | Crystal structure of l-fuculose-1-phosphate aldolase from Thermus Thermophilus HB8 | | Descriptor: | fuculose-1-phosphate aldolase | | Authors: | Jeyakanthan, J, Yokoyama, S, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-06 | | Release date: | 2007-01-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Purification, crystallization and preliminary X-ray crystallographic study of the L-fuculose-1-phosphate aldolase (FucA) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1UJN
 
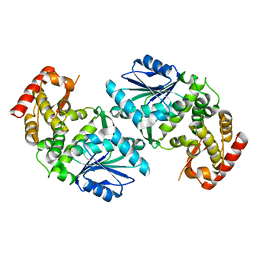 | | Crystal structure of dehydroquinate synthase from Thermus thermophilus HB8 | | Descriptor: | dehydroquinate synthase | | Authors: | Sugahara, M, Yokoyama, S, Kuramitsu, S, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-06 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of dehydroquinate synthase from Thermus thermophilus HB8 showing functional importance of the dimeric state.
Proteins, 58, 2005
|
|
1UKK
 
 | | Structure of Osmotically Inducible Protein C from Thermus thermophilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Osmotically Inducible Protein C | | Authors: | Rehse, P.H, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Structure and Biochemical Analysis of the Thermus thermophilus Osmotically Inducible Protein C
J.MOL.BIOL., 338, 2004
|
|
4YN3
 
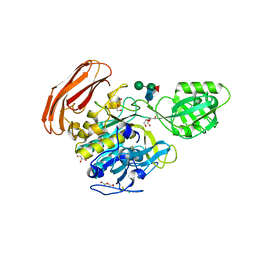 | | Crystal structure of Cucumisin complex with pro-peptide | | Descriptor: | CHLORIDE ION, Cucumisin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murayama, K, Kato-Murayama, M, Yokoyama, S, Arima, K, Shirouzu, M. | | Deposit date: | 2015-03-09 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of cucumisin protease activity regulation by its propeptide
J. Biochem., 161, 2017
|
|
3ACD
 
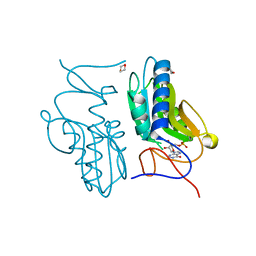 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase with IMP from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Hypoxanthine-guanine phosphoribosyltransferase, INOSINIC ACID | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3ACC
 
 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase with GMP from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3ACB
 
 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
