2MGZ
 
 | | Solution structure of RBFOX family ASD-1 RRM and SUP-12 RRM in ternary complex with RNA | | Descriptor: | Protein ASD-1, isoform a, Protein SUP-12, ... | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
2ZET
 
 | | Crystal structure of the small GTPase Rab27B complexed with the Slp homology domain of Slac2-a/melanophilin | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Melanophilin, ... | | Authors: | Kukimoto-Niino, M, Sakamoto, A, Kanno, E, Hanawa-Suetsugu, K, Terada, T, Shirouzu, M, Fukuda, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-12-17 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases.
Structure, 16, 2008
|
|
2Z34
 
 | |
2PBR
 
 | | Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5 | | Descriptor: | SULFATE ION, Thymidylate kinase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5
To be Published
|
|
3AQA
 
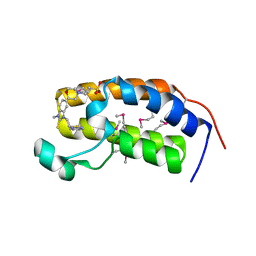 | | Crystal structure of the human BRD2 BD1 bromodomain in complex with a BRD2-interactive compound, BIC1 | | Descriptor: | 1-[2-(1H-benzimidazol-2-ylsulfanyl)ethyl]-3-methyl-1,3-dihydro-2H-benzimidazole-2-thione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Umehara, T, Nakamura, Y, Terada, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-10-27 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Real-Time Imaging of Histone H4K12-Specific Acetylation Determines the Modes of Action of Histone Deacetylase and Bromodomain Inhibitors
Chem.Biol., 18, 2011
|
|
3AY9
 
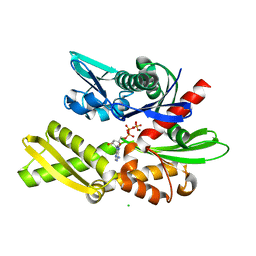 | | Crystal structure of human Hsp70 NBD in the ADP-, Mg ion-, and K ion-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Arakawa, A, Handa, N, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-05-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural studies on the high affinity of Hsp70 for ADP.
Protein Sci., 20, 2011
|
|
2YR1
 
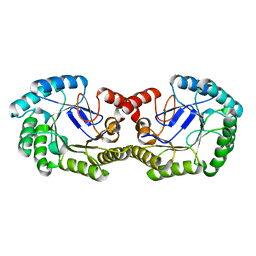 | | Crystal Structure of 3-dehydroquinate dehydratase from Geobacillus kaustophilus HTA426 | | Descriptor: | 3-dehydroquinate dehydratase | | Authors: | Kagawa, W, Kurumizaka, H, Bessho, Y, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-01 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of 3-dehydroquinate dehydratase from Geobacillus kaustophilus HTA426
To be published
|
|
2YWH
 
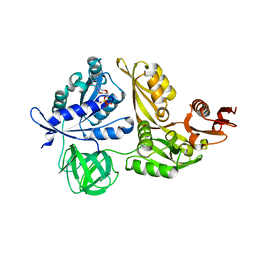 | | Crystal structure of GDP-bound LepA from Aquifex aeolicus | | Descriptor: | GTP-binding protein LepA, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Nishino, A, Nakayama-Ushikoshi, R, Hanawa-Suetsugu, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of GTP-binding protein LepA from Aquifex aeolicus.
To be Published
|
|
2YUH
 
 | |
1WFD
 
 | |
2YV5
 
 | | Crystal structure of Yjeq from Aquifex aeolicus | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, YjeQ protein, ... | | Authors: | Wang, H, Kaminishi, T, Hanawa-Suetsugu, K, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-09 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of YjeQ from Aquifex aeolicus
To be Published
|
|
1WEP
 
 | | Solution structure of PHD domain in PHF8 | | Descriptor: | PHF8, ZINC ION | | Authors: | He, F, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in protein AA017385
To be Published
|
|
1WF5
 
 | |
2YVU
 
 | |
1WI4
 
 | | Solution structure of the PDZ domain of syntaxin binding protein 4 | | Descriptor: | syntaxin binding protein 4 | | Authors: | Endo, H, Tomizawa, T, Kigawa, T, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of syntaxin binding protein 4
To be published
|
|
2YR3
 
 | | Solution structure of the fourth Ig-like domain from myosin light chain kinase, smooth muscle | | Descriptor: | Myosin light chain kinase, smooth muscle | | Authors: | Qin, X.R, Kurosaki, C, Yoshida, M, Hayahsi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth Ig-like domain from myosin light chain kinase, smooth muscle
To be Published
|
|
1WJ0
 
 | |
2YT8
 
 | | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma) | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 3 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Seiki, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma)
To be Published
|
|
2PBP
 
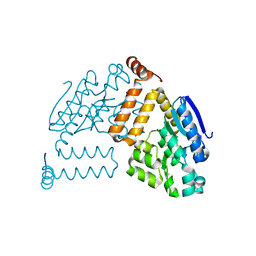 | | Crystal structure of ENOYL-CoA hydrates subunit I (gk_2039) from geobacillus kaustophilus HTA426 | | Descriptor: | Enoyl-CoA hydratase subunit I | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki, R.C, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ENOYL-CoA hydrates subunit I (gk_2039) from geobacillus kaustophilus HTA426
To be Published
|
|
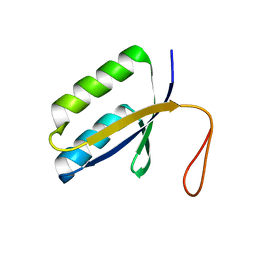
1ZPW
 
 | |
1X49
 
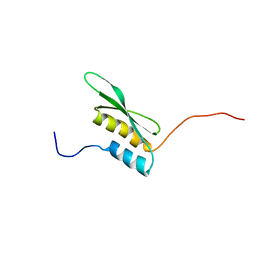 | | Solution structure of the first DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase
To be Published
|
|
1X4N
 
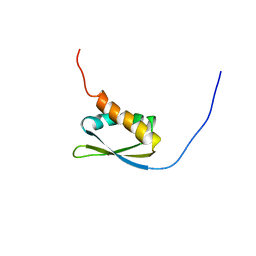 | | Solution structure of KH domain in FUSE binding protein 1 | | Descriptor: | Far upstream element binding protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of KH domain in FUSE binding protein 1
To be Published
|
|
1X5R
 
 | |
1X4R
 
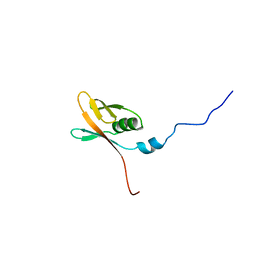 | | Solution structure of WWE domain in Parp14 protein | | Descriptor: | Parp14 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of WWE domain in Parp14 protein
To be Published
|
|
2PE3
 
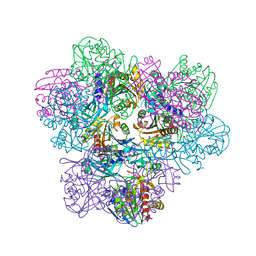 | | Crystal structure of Frv operon protein FRVX (PH1821)from pyrococcus horikoshii OT3 | | Descriptor: | 354aa long hypothetical operon protein Frv | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Rafi, Z.A, Sekar, K, Inagakai, E, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of frv operon protein frvx (ph1821)from pyrococcus horikoshii OT3
To be Published
|
|
