4X39
 
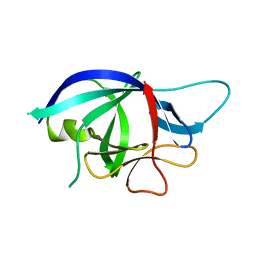 | |
4X38
 
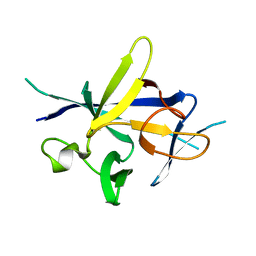 | |
4X37
 
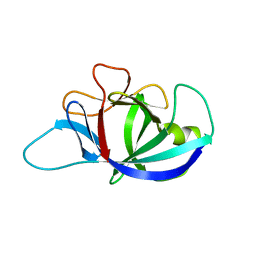 | |
4X3A
 
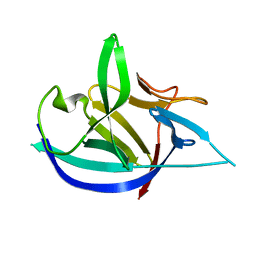 | |
1ZTM
 
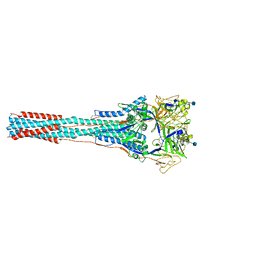 | | Structure of the Uncleaved Paramyxovirus (hPIV3) Fusion Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein | | Authors: | Yin, H.S, Paterson, R.G, Wen, X, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-05-27 | | Release date: | 2005-07-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3NJ5
 
 | |
3OTW
 
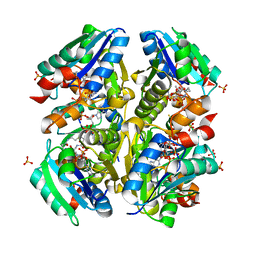 | | Structural and Functional Studies of Helicobacter pylori Wild-Type and Mutated Proteins Phosphopantetheine adenylyltransferase | | Descriptor: | COENZYME A, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Yin, H.S, Cheng, C.S, Chen, C.G, Luo, Y.C, Chen, W.T, Cheng, S.Y. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Studies of Helicobacter pylori Wild-Type and Mutated Proteins Phosphopantetheine adenylyltransferase
To be Published
|
|
7CPR
 
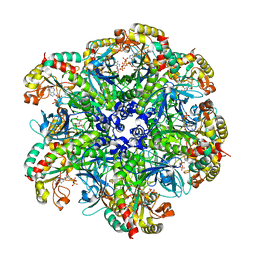 | | glutamine synthetase from Drosophila | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase 2 cytoplasmic | | Authors: | Yin, H.S, Chen, W.T. | | Deposit date: | 2020-08-07 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Insight into the Contributions of the N-Terminus and Key Active-Site Residues to the Catalytic Efficiency of Glutamine Synthetase 2.
Biomolecules, 10, 2020
|
|
8JA0
 
 | |
5GZM
 
 | | Cyclodeaminase_PA | | Descriptor: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ying, H, Chen, K. | | Deposit date: | 2016-09-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclodeaminase_PA
To Be Published
|
|
5GZL
 
 | | Cyclodeaminase_PA | | Descriptor: | LYSINE, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ying, H, Chen, K. | | Deposit date: | 2016-09-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclodeaminase_PA
To Be Published
|
|
5GZI
 
 | | Cyclodeaminase_PA | | Descriptor: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ying, H, Chen, K. | | Deposit date: | 2016-09-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Cyclodeaminase_PA
To Be Published
|
|
5GZJ
 
 | | Cyclodeaminase_PA | | Descriptor: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ying, H, Chen, K. | | Deposit date: | 2016-09-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Cyclodeaminase_PA
To Be Published
|
|
2WRY
 
 | | Crystal structure of chicken cytokine interleukin 1 beta | | Descriptor: | INTERLEUKIN-1BETA | | Authors: | Lu, W.S, Cheng, C.S, Lyu, P.C, Lee, L.H, Wang, W.C, Yin, H.S. | | Deposit date: | 2009-09-03 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Functional Comparison of Cytokine Interleukin-1 Beta from Chicken and Human.
Mol.Immunol., 48, 2011
|
|
5WSZ
 
 | | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis | | Descriptor: | COPPER (II) ION, LpmO10A | | Authors: | Zhao, Y, Zhang, H, Yin, H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis
To Be Published
|
|
3NV7
 
 | |
3RVG
 
 | | Crystals structure of Jak2 with a 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitor | | Descriptor: | 1-(cyclohexylamino)-7-(1-methyl-1H-pyrazol-4-yl)-5H-pyrido[4,3-b]indole-4-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Lim, J, Taoka, B, Otte, R.D, Spencer, K, Dinsmore, C.J, Altman, M.D, Chan, G, Rosenstein, C, Sharma, S, Su, H.P, Szewczak, A.A, Xu, L, Yin, H, Zugay-Murphy, J, Marshall, C.G, Young, J.R. | | Deposit date: | 2011-05-06 | | Release date: | 2012-03-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Discovery of 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitors of Janus kinase 2 (JAK2) for the treatment of myeloproliferative disorders.
J.Med.Chem., 54, 2011
|
|
8IYS
 
 | | TUG891-bound FFAR4 in complex with Gq | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4I
 
 | | DHA-bound FFAR4 in complex with Gs | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4L
 
 | | DHA-bound FFAR4 in complex with Gq | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4K
 
 | | GW9508-bound FFAR4 in complex with Gq | | Descriptor: | 3-(4-{[(3-phenoxyphenyl)methyl]amino}phenyl)propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
7X3Z
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 1 complexed with estrone and NAD | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, 17-beta-hydroxysteroid dehydrogenase type 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, T, Lin, S.X, Yin, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New insights into the substrate inhibition of human 17 beta-hydroxysteroid dehydrogenase type 1.
J.Steroid Biochem.Mol.Biol., 228, 2023
|
|
7WJQ
 
 | | Crystal structure of GSDMB in complex with Ipah7.8 | | Descriptor: | Isoform 2 of Gasdermin-B, Probable E3 ubiquitin-protein ligase ipaH7.8 | | Authors: | Li, X, Zhang, H, Yin, H. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the GSDMB-mediated cellular lysis and its targeting by IpaH7.8
Nat Commun, 14, 2023
|
|
7V8O
 
 | | Crystal structure of cyclohexanone monooxygenase from T. municipale mutant L437T complexed with NADP+ and FAD in space group of P21221 | | Descriptor: | 1,2-ETHANEDIOL, Cyclohexanone Monooxygenase from Thermocrispum municipale, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, T, Li, G.Y, Yin, H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Biocatalytic Baeyer-Villiger Reactions: Uncovering the Source of Regioselectivity at Each Evolutionary Stage of a Mutant with Scrutiny of Fleeting Chiral Intermediates.
Acs Catalysis, 12, 2022
|
|
7V8R
 
 | | Crystal structure of cyclohexanone monooxygenase from T. municipale mutant L437T complexed with NADP+ and FAD in space group of C2221 | | Descriptor: | 1,2-ETHANEDIOL, Cyclohexanone Monooxygenase from Thermocrispum municipale, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, T, Li, G.Y, Yin, H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Biocatalytic Baeyer-Villiger Reactions: Uncovering the Source of Regioselectivity at Each Evolutionary Stage of a Mutant with Scrutiny of Fleeting Chiral Intermediates.
Acs Catalysis, 12, 2022
|
|
