5YXJ
 
 | | FXR ligand binding domain | | Descriptor: | 2-[benzyl(methyl)amino]ethyl methyl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Yi, L, Yong, L. | | Deposit date: | 2017-12-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A ligand of drug binding to FXR
To Be Published
|
|
5YXD
 
 | | A ligand F binding to FXR | | Descriptor: | Bile acid receptor, Peptide from Nuclear receptor coactivator, ethyl methyl 4-(2,3-dichlorophenyl)-2,6-dimethylpyridine-3,5-dicarboxylate | | Authors: | Yi, L, Yong, L. | | Deposit date: | 2017-12-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A ligand F binding to FXR
To Be Published
|
|
5YXL
 
 | | A ligand M binding to FXR | | Descriptor: | 2-methoxyethyl propan-2-yl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Yi, L, Yong, L. | | Deposit date: | 2017-12-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A ligand M binding to FXR
To Be Published
|
|
5YXB
 
 | | A ligand binding to FXR | | Descriptor: | 2-methoxyethyl (2E)-3-phenylprop-2-en-1-yl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Yi, L, Yong, L. | | Deposit date: | 2017-12-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A ligand binding to FXR
To Be Published
|
|
5YXU
 
 | |
5SYD
 
 | |
3ELO
 
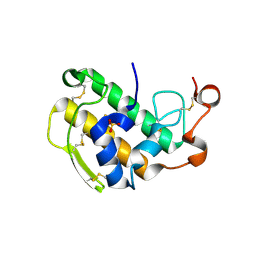 | | Crystal Structure of Human Pancreatic Prophospholipase A2 | | Descriptor: | Phospholipase A2, SULFATE ION | | Authors: | Xu, W, Yi, L, Feng, Y, Chen, L, Liu, J. | | Deposit date: | 2008-09-22 | | Release date: | 2009-04-14 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into the activation mechanism of human pancreatic prophospholipase A2
J.Biol.Chem., 284, 2009
|
|
4LCT
 
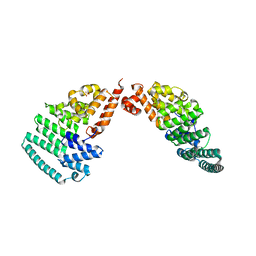 | | Crystal Structure and Versatile Functional Roles of the COP9 Signalosome Subunit 1 | | Descriptor: | COP9 signalosome complex subunit 1, SULFATE ION | | Authors: | Lee, J.-H, Yi, L, Li, J, Schweitzer, K, Borgmann, M, Naumann, M, Wu, H. | | Deposit date: | 2013-06-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and versatile functional roles of the COP9 signalosome subunit 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5ZG9
 
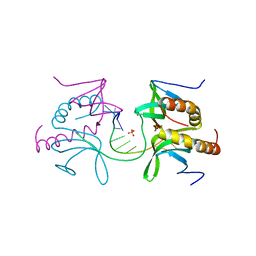 | | Crystal structure of MoSub1-ssDNA complex in phosphate buffer | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*G)-3'), MoSub1, PHOSPHATE ION | | Authors: | Zhao, Y, Huang, J, Liu, H, Yi, L, Wang, S, Zhang, X, Liu, J. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The effect of phosphate ion on the ssDNA binding mode of MoSub1, a Sub1/PC4 homolog from rice blast fungus.
Proteins, 87, 2019
|
|
6JBI
 
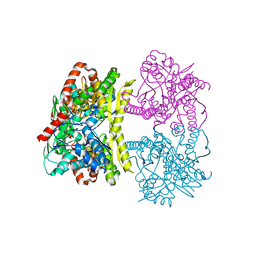 | | Structure of Tps1 apo structure | | Descriptor: | Trehalose-6-phosphate synthase | | Authors: | Wang, S, Zhao, Y, Yi, L, Wang, D, Liu, J. | | Deposit date: | 2019-01-25 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
