7AY7
 
 | | Structure of SARS-CoV-2 Main Protease bound to Isofloxythepin | | Descriptor: | 3C-like proteinase, 9-fluoranyl-3-propan-2-yl-5,6-dihydrobenzo[b][1]benzothiepine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWR
 
 | | Structure of SARS-CoV-2 Main Protease bound to Tegafur | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, TEGAFUR | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AX6
 
 | | Structure of SARS-CoV-2 Main Protease bound to Glutathione isopropyl ester | | Descriptor: | (2~{S})-2-azanyl-5-oxidanylidene-5-[[(2~{S})-1-oxidanylidene-1-[(2-oxidanylidene-2-propan-2-yloxy-ethyl)amino]-3-sulfanyl-propan-2-yl]amino]pentanoic acid, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AR6
 
 | | Structure of apo SARS-CoV-2 Main Protease with large beta angle, space group C2. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AP6
 
 | | Structure of SARS-CoV-2 Main Protease bound to MUT056399. | | Descriptor: | 3C-like proteinase, 4-(4-ethyl-5-fluoranyl-2-oxidanyl-phenoxy)-3-fluoranyl-benzamide | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ARF
 
 | | Structure of SARS-CoV-2 Main Protease bound to thioglucose. | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-sulfanyl-oxane-3,4,5-triol, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWS
 
 | | Structure of SARS-CoV-2 Main Protease bound to TH-302. | | Descriptor: | 3C-like proteinase, 5-[[(2-bromoethylamino)-(ethylamino)phosphoryl]oxymethyl]-1-methyl-~{N},~{N}-bis(oxidanyl)imidazol-2-amine, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AXM
 
 | | Structure of SARS-CoV-2 Main Protease bound to Pelitinib | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-3-cyano-7-ethoxyquinolin-6-yl}-4-(dimethylamino)but-2-enamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8BEY
 
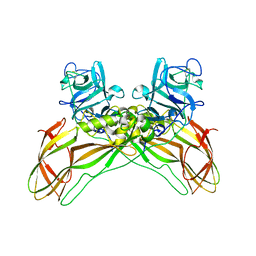 | | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein at pH 7 | | Descriptor: | Cry49Aa protein | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R. | | Deposit date: | 2022-10-22 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BEX
 
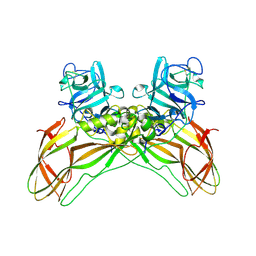 | | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein at pH 3 | | Descriptor: | Cry49Aa protein | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R. | | Deposit date: | 2022-10-22 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BEZ
 
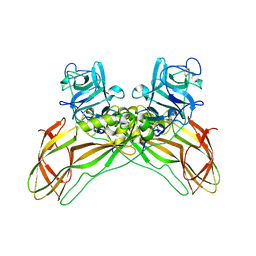 | | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein at pH 11 | | Descriptor: | Cry49Aa protein | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R. | | Deposit date: | 2022-10-22 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7B83
 
 | | Structure of SARS-CoV-2 Main Protease bound to pyrithione zinc | | Descriptor: | 3C-like proteinase, 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8PCF
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 500 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCS
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 1250 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCI
 
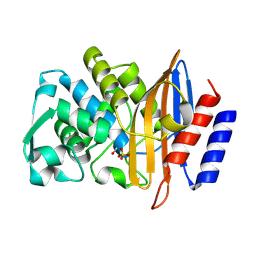 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 5000 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCG
 
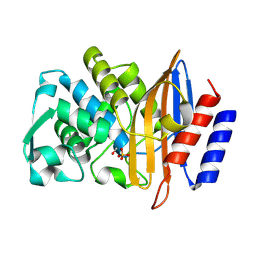 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 2000 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCK
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 0 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCN
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 100 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PC9
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 0 ms - native. | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCL
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 50 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCQ
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 500 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCC
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 100 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCV
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 10000 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCB
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 80 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCO
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 150 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
