7AK4
 
 | | Structure of SARS-CoV-2 Main Protease bound to Tretazicar. | | Descriptor: | 3C-like proteinase, 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AKU
 
 | | Structure of SARS-CoV-2 Main Protease bound to Calpeptin. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, Calpeptin | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-02 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ARF
 
 | | Structure of SARS-CoV-2 Main Protease bound to thioglucose. | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-sulfanyl-oxane-3,4,5-triol, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWR
 
 | | Structure of SARS-CoV-2 Main Protease bound to Tegafur | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, TEGAFUR | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AX6
 
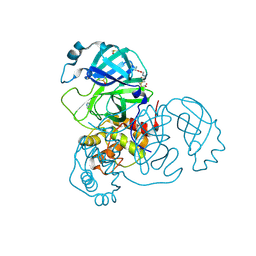 | | Structure of SARS-CoV-2 Main Protease bound to Glutathione isopropyl ester | | Descriptor: | (2~{S})-2-azanyl-5-oxidanylidene-5-[[(2~{S})-1-oxidanylidene-1-[(2-oxidanylidene-2-propan-2-yloxy-ethyl)amino]-3-sulfanyl-propan-2-yl]amino]pentanoic acid, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AR6
 
 | | Structure of apo SARS-CoV-2 Main Protease with large beta angle, space group C2. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AY7
 
 | | Structure of SARS-CoV-2 Main Protease bound to Isofloxythepin | | Descriptor: | 3C-like proteinase, 9-fluoranyl-3-propan-2-yl-5,6-dihydrobenzo[b][1]benzothiepine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWS
 
 | | Structure of SARS-CoV-2 Main Protease bound to TH-302. | | Descriptor: | 3C-like proteinase, 5-[[(2-bromoethylamino)-(ethylamino)phosphoryl]oxymethyl]-1-methyl-~{N},~{N}-bis(oxidanyl)imidazol-2-amine, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AXM
 
 | | Structure of SARS-CoV-2 Main Protease bound to Pelitinib | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-3-cyano-7-ethoxyquinolin-6-yl}-4-(dimethylamino)but-2-enamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6QF1
 
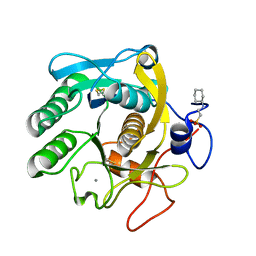 | | X-Ray structure of Proteinase K crystallized on a silicon chip | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Proteinase K | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QF2
 
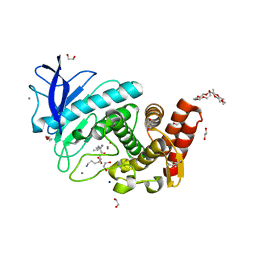 | | X-Ray structure of Thermolysin crystallized on a silicon chip | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CALCIUM ION, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QF3
 
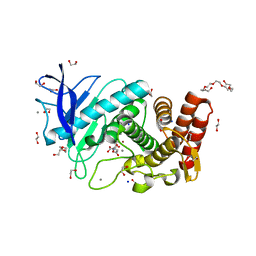 | | X-Ray structure of Thermolysin soaked with sodium aspartate on a silicon chip | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ASPARTIC ACID, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QF4
 
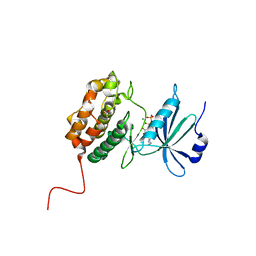 | | X-Ray structure of human Serine/Threonine Kinase 17B (STK17B) aka DRAK2 in complex with ADP obtained by on-chip soaking | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QF5
 
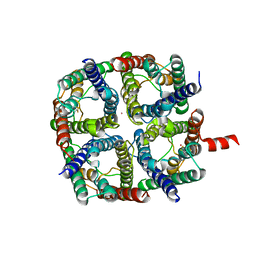 | | X-Ray structure of human Aquaporin 2 crystallized on a silicon chip | | Descriptor: | Aquaporin-2, CADMIUM ION | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
7AVD
 
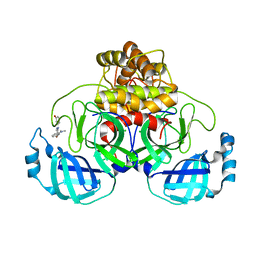 | | Structure of SARS-CoV-2 Main Protease bound to SEN1269 ligand | | Descriptor: | 3-[[5-[3-(dimethylamino)phenoxy]pyrimidin-2-yl]amino]phenol, 3C-like proteinase, CHLORIDE ION | | Authors: | Koua, F, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Ewert, W, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AQJ
 
 | | Structure of SARS-CoV-2 Main Protease bound to Triglycidyl isocyanurate. | | Descriptor: | 1-[(2~{R})-2-oxidanylpropyl]-3-[[(2~{R})-oxiran-2-yl]methyl]-5-[[(2~{S})-oxiran-2-yl]methyl]-1,3,5-triazinane-2,4,6-trione, 3C-like proteinase, Triglycidyl isocyanurate | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AP6
 
 | | Structure of SARS-CoV-2 Main Protease bound to MUT056399. | | Descriptor: | 3C-like proteinase, 4-(4-ethyl-5-fluoranyl-2-oxidanyl-phenoxy)-3-fluoranyl-benzamide | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7B83
 
 | | Structure of SARS-CoV-2 Main Protease bound to pyrithione zinc | | Descriptor: | 3C-like proteinase, 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8OY6
 
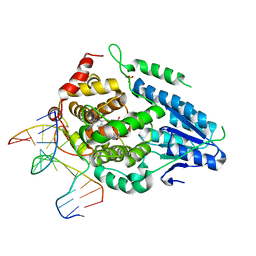 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (3 nanosecond pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OYA
 
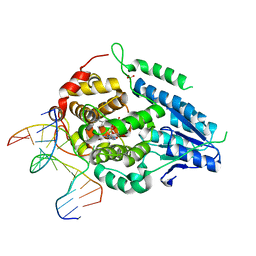 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (10 microsecond pump probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OET
 
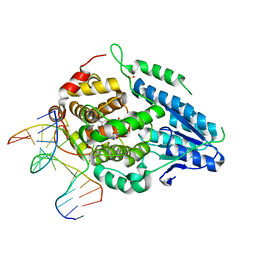 | | SFX structure of the class II photolyase complexed with a thymine dimer | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, DNA (14-mer), Deoxyribodipyrimidine photo-lyase, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-03-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OYC
 
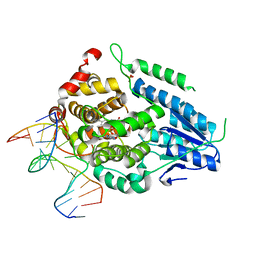 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (100 microsecond timpeoint) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OY8
 
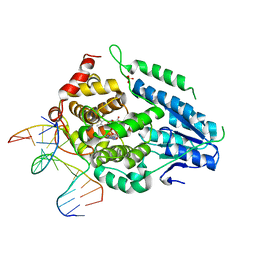 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (30 nanosecond timepoint) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OY3
 
 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (3 picosecond pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OY5
 
 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (1 nanosecond pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
