5IC7
 
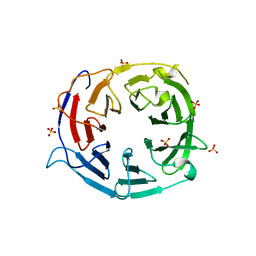 | | Structure of the WD domain of UTP18 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-22 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.331 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5IC9
 
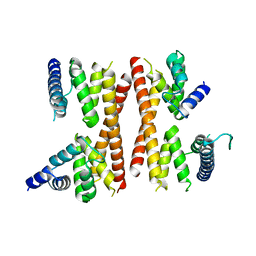 | | Structure of the CTD complex of Utp12 and Utp13 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5IC8
 
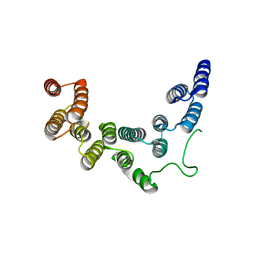 | | Structure of UTP6 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
4M5D
 
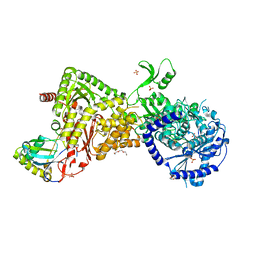 | |
4J0X
 
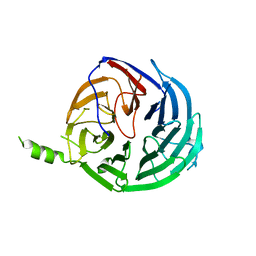 | | Structure of Rrp9 | | Descriptor: | Ribosomal RNA-processing protein 9 | | Authors: | Zhang, L, Lin, J, Ye, K. | | Deposit date: | 2013-01-31 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural and functional analysis of the U3 snoRNA binding protein Rrp9.
Rna, 19, 2013
|
|
4J0W
 
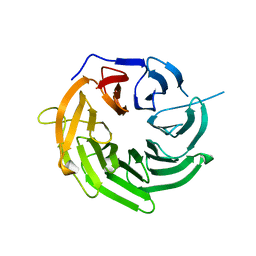 | | Structure of U3-55K | | Descriptor: | U3 small nucleolar RNA-interacting protein 2 | | Authors: | Zhang, L, Lin, J, Ye, K. | | Deposit date: | 2013-01-31 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional analysis of the U3 snoRNA binding protein Rrp9.
Rna, 19, 2013
|
|
4DJG
 
 | |
4DOZ
 
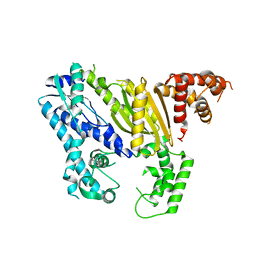 | | Crystal structure of Pyrococcus furiosus Cmr2 (Cas10) | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Zhu, X, Ye, K. | | Deposit date: | 2012-02-12 | | Release date: | 2012-03-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR-Cas systems
Febs Lett., 586, 2012
|
|
6KE6
 
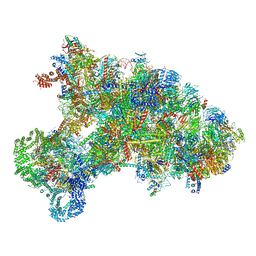 | |
5YDU
 
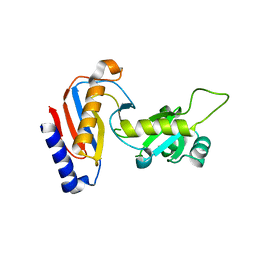 | | Crystal structure of Utp30 | | Descriptor: | PHOSPHATE ION, Ribosome biogenesis protein UTP30 | | Authors: | Hu, J, Zhu, X, Ye, K. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
7VQO
 
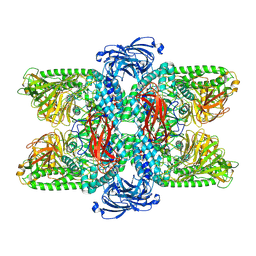 | | Cryo-EM structure of Ams1 bound to the FW domain of Nbr1 | | Descriptor: | Ams1, Nbr1 and malE fusion protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural mechanism of protein recognition by the FW domain of autophagy receptor Nbr1
Nat Commun, 13, 2022
|
|
5YMA
 
 | | Crystal structure of ribosome assembly factor Efg1 | | Descriptor: | Putative rRNA processing protein | | Authors: | Shu, S, Ye, K. | | Deposit date: | 2017-10-21 | | Release date: | 2018-01-17 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Structural and functional analysis of ribosome assembly factor Efg1.
Nucleic Acids Res., 46, 2018
|
|
5YZ4
 
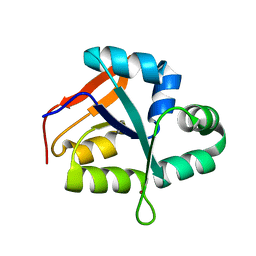 | | Structure of the PIN domain endonuclease Utp24 | | Descriptor: | CALCIUM ION, ZINC ION, rRNA-processing protein fcf1 | | Authors: | Du, Y, An, W, Ye, K. | | Deposit date: | 2017-12-12 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.135 Å) | | Cite: | Structural and functional analysis of Utp24, an endonuclease for processing 18S ribosomal RNA.
Plos One, 13, 2018
|
|
5ZEW
 
 | |
5Z1G
 
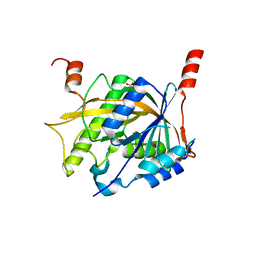 | | Structure of the Brx1 and Ebp2 complex | | Descriptor: | Ribosome biogenesis protein BRX1, SULFATE ION, rRNA-processing protein EBP2 | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2017-12-26 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate
Protein Cell, 10, 2019
|
|
5WY3
 
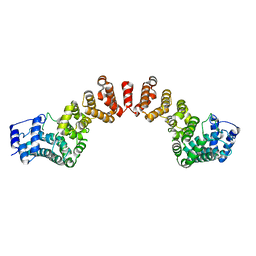 | |
6IF4
 
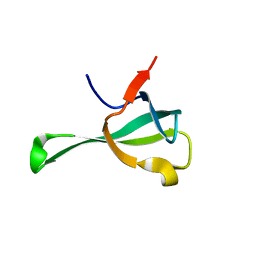 | | Crystal structure of Tbtudor | | Descriptor: | Histone acetyltransferase | | Authors: | Gao, J, Ye, K, Diwu, Y, Liao, S, Tu, X. | | Deposit date: | 2018-09-18 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.934 Å) | | Cite: | Crystal structure of TbEsa1 presumed Tudor domain from Trypanosoma brucei.
J.Struct.Biol., 209, 2020
|
|
7DDE
 
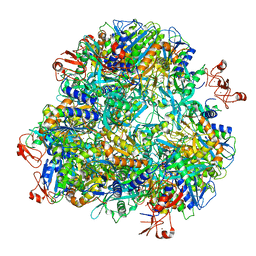 | | Cryo-EM structure of the Ape4 and Nbr1 complex | | Descriptor: | Aspartyl aminopeptidase 1,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
7D4I
 
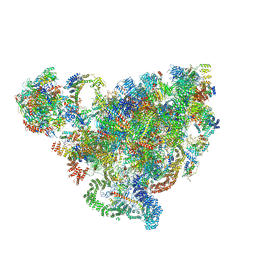 | | Cryo-EM structure of 90S small ribosomal precursors complex with the DEAH-box RNA helicase Dhr1 (State F) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of 90S small ribosomal precursors complex with Dhr1
To Be Published
|
|
7D5S
 
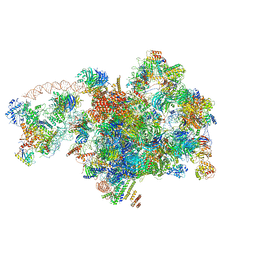 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S12, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2)
To Be Published
|
|
7D63
 
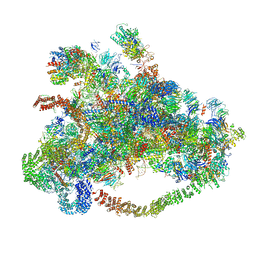 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C)
To Be Published
|
|
7D5T
 
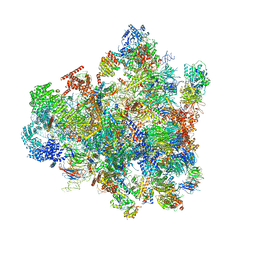 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1)
To Be Published
|
|
7DD9
 
 | | Cryo-EM structure of the Ams1 and Nbr1 complex | | Descriptor: | Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
2F3W
 
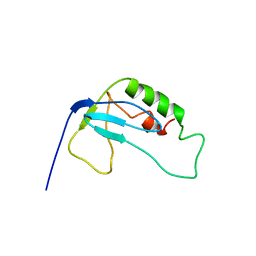 | | solution structure of 1-110 fragment of staphylococcal nuclease in 2M TMAO | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
2F3V
 
 | | Solution structure of 1-110 fragment of staphylococcal nuclease with V66W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
