8H1L
 
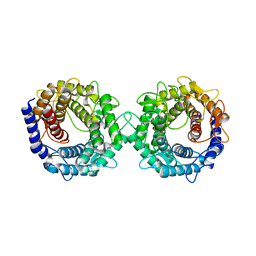 | | Crystal structure of glucose-2-epimerase in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1M
 
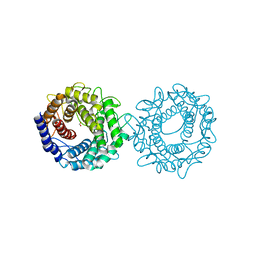 | | Crystal structure of glucose-2-epimerase mutant_D254A from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1N
 
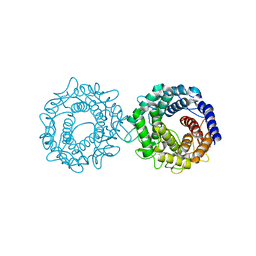 | | Crystal structure of glucose-2-epimerase mutant_D254A in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
2ZCX
 
 | |
2ZPA
 
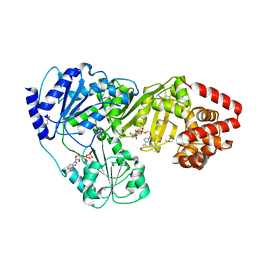 | | Crystal Structure of tRNA(Met) Cytidine Acetyltransferase | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Chimnaronk, S, Manita, T, Yao, M, Tanaka, I. | | Deposit date: | 2008-07-08 | | Release date: | 2009-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon
Embo J., 28, 2009
|
|
2ZDS
 
 | | Crystal Structure of SCO6571 from Streptomyces coelicolor A3(2) | | Descriptor: | Putative DNA-binding protein | | Authors: | Begum, P, Gao, Y.G, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-02 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SCO6571 from Streptomyces coelicolor A3(2).
Protein Pept.Lett., 15, 2008
|
|
8IN4
 
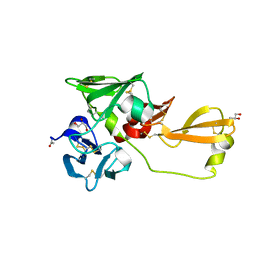 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, ACETYL GROUP, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN1
 
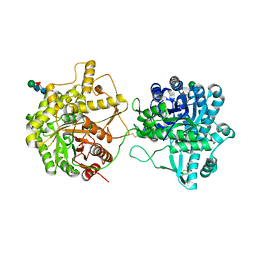 | | beta-glucosidase protein from Aplysia kurodai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN3
 
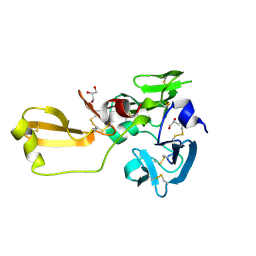 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN6
 
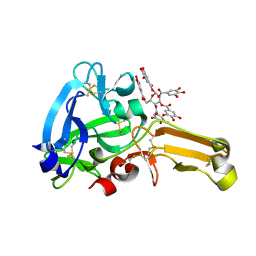 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai complex with tannic acid | | Descriptor: | 25 kDa polyphenol-binding protein, BETA-1,2,3,4,6-PENTA-O-GALLOYL-D-GLUCOPYRANOSE | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
3A7W
 
 | |
3A88
 
 | |
3A7V
 
 | |
3A87
 
 | |
3A7T
 
 | |
3A83
 
 | |
3A82
 
 | |
3A8D
 
 | |
2ZB9
 
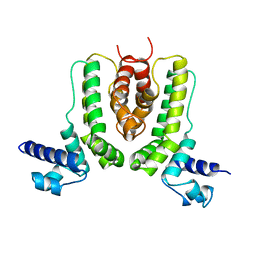 | | Crystal structure of TetR family transcription regulator SCO0332 | | Descriptor: | Putative transcriptional regulator | | Authors: | Okada, U, Kondo, K, Watanabe, N, Yao, M, Tanaka, I. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional analysis of the TetR-family transcriptional regulator SCO0332 from Streptomyces coelicolor
ACTA CRYSTALLOGR.,SECT.D, 64, 2008
|
|
3A89
 
 | |
3A84
 
 | |
3ANZ
 
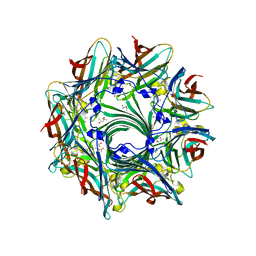 | | Crystal Structure of alpha-hemolysin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Alpha-hemolysin | | Authors: | Yamashita, K, Kawauchi, H, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2010-09-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal alpha-hemolysin into heptameric pore structure
Protein Sci., 20, 2011
|
|
3A7Z
 
 | |
3A8B
 
 | |
3A81
 
 | |
