1N03
 
 | | Model for Active RecA Filament | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, RecA protein | | 著者 | VanLoock, M.S, Yu, X, Yang, S, Lai, A.L, Low, C, Campbell, M.J, Egelman, E.H. | | 登録日 | 2002-10-10 | | 公開日 | 2003-02-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | ATP-Mediated Conformational Changes in the RecA Filament
Structure, 11, 2003
|
|
8TID
 
 | | Combined linker domain of N-DRC and associated proteins Tetrahymena | | 分子名称: | AAA family ATPase CDC48 subfamily protein, CFAP20, Calmodulin 7-2, ... | | 著者 | Ghanaeian, A.G, Majhi, S.M, McCaffrey, C.M, Nami, B.N, Black, C.B, Yang, S.K, Legal, T.L, Papoulas, O.P, Janowska, M.J, Valente-Paterno, M.V, Marcotte, E.M, Wloga, D.W, Bui, K.H. | | 登録日 | 2023-07-19 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Integrated modeling of the Nexin-dynein regulatory complex reveals its regulatory mechanism.
Nat Commun, 14, 2023
|
|
3JBM
 
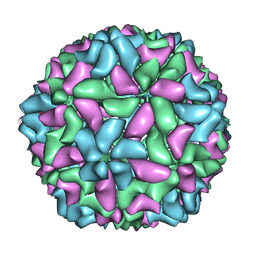 | | Electron cryo-microscopy of a virus-like particle of orange-spotted grouper nervous necrosis virus | | 分子名称: | virus-like particle of orange-spotted grouper nervous necrosis virus | | 著者 | Xie, J, Li, K, Gao, Y, Huang, R, Lai, Y, Shi, Y, Yang, S, Zhu, G, Zhang, Q, He, J. | | 登録日 | 2015-09-06 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle
Vet. Res., 47, 2016
|
|
1QN4
 
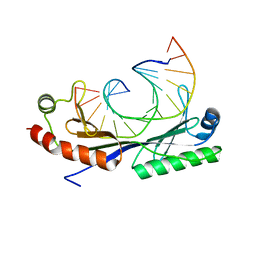 | | Crystal structure of the T(-24) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | 分子名称: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*TP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*AP*TP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | 著者 | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | 登録日 | 1999-10-14 | | 公開日 | 2000-02-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QNB
 
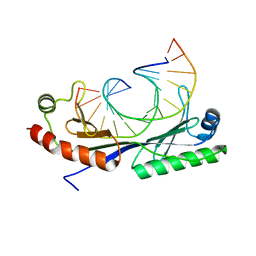 | | Crystal structure of the T(-25) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | 分子名称: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*TP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*AP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | 著者 | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | 登録日 | 1999-10-14 | | 公開日 | 2000-02-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN8
 
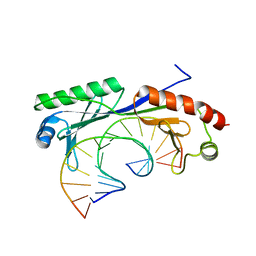 | | Crystal structure of the T(-28) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | 分子名称: | DNA (5'-D(*GP*CP*TP*AP*TP*TP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*AP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | 著者 | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | 登録日 | 1999-10-14 | | 公開日 | 2000-02-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QNC
 
 | | Crystal structure of the A(-31) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | 分子名称: | DNA (5'-D(*GP*CP*AP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*TP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | 著者 | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | 登録日 | 1999-10-14 | | 公開日 | 2000-02-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN6
 
 | | Crystal structure of the T(-26) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | 分子名称: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*AP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | 著者 | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | 登録日 | 1999-10-14 | | 公開日 | 2000-02-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN9
 
 | | Crystal structure of the C(-29) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | 分子名称: | DNA (5'-D(*GP*CP*TP*AP*CP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*GP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | 著者 | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | 登録日 | 1999-10-14 | | 公開日 | 2000-02-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QNA
 
 | | Crystal structure of the T(-30) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | 分子名称: | DNA (5'-D(*GP*CP*TP*TP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*AP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | 著者 | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | 登録日 | 1999-10-14 | | 公開日 | 2000-02-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN3
 
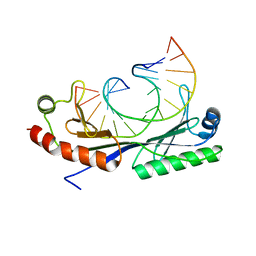 | | Crystal structure of the C(-25) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | 分子名称: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*CP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*GP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | 著者 | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | 登録日 | 1999-10-14 | | 公開日 | 2000-02-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
8SF7
 
 | | 48-nm doublet microtubule from Tetrahymena thermophila strain MEC17 | | 分子名称: | CFAM166A, CFAM166B, CFAM166C, ... | | 著者 | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | 登録日 | 2023-04-10 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Effect of alpha-tubulin acetylation on the doublet microtubule structure.
Elife, 12, 2024
|
|
8TEK
 
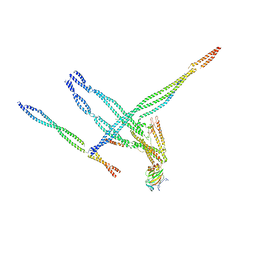 | | Baseplate of Nexin-dynein regulatory complex from Tetrahymena thermophila | | 分子名称: | CFAP20, Cilia- and flagella-associated protein 91, Coiled-coil protein, ... | | 著者 | Ghanaeian, A.G, Black, C.S, Yang, S.K, Bui, K.H. | | 登録日 | 2023-07-06 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Integrated modeling of the Nexin-dynein regulatory complex reveals its regulatory mechanism.
Nat Commun, 14, 2023
|
|
1QN7
 
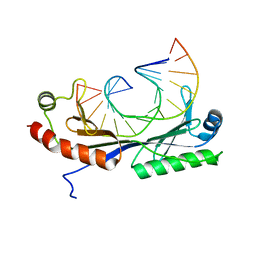 | | Crystal structure of the T(-27) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | 分子名称: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*TP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*AP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | 著者 | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | 登録日 | 1999-10-14 | | 公開日 | 2000-02-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN5
 
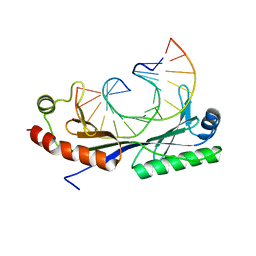 | | Crystal structure of the G(-26) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | 分子名称: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*GP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*CP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | 著者 | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | 登録日 | 1999-10-14 | | 公開日 | 2000-02-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
8T6V
 
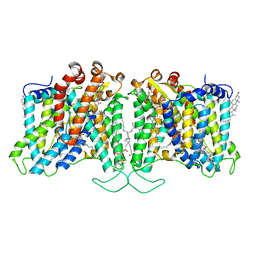 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (DIDS) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | 著者 | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2023-06-18 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T6U
 
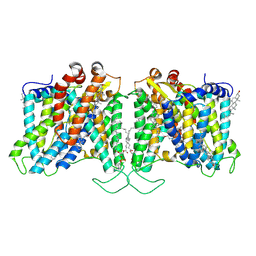 | | Cryo-EM structure of human Anion Exchanger 1 bound to Dipyridamole | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2023-06-18 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1VYT
 
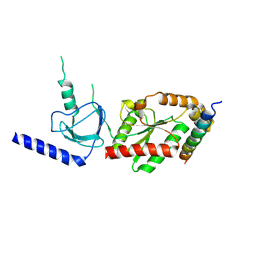 | | beta3 subunit complexed with aid | | 分子名称: | CALCIUM CHANNEL BETA-3 SUBUNIT, VOLTAGE-DEPENDENT L-TYPE CALCIUM CHANNEL ALPHA-1C SUBUNIT | | 著者 | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Tong, L, Yang, J. | | 登録日 | 2004-05-07 | | 公開日 | 2004-06-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
6JFX
 
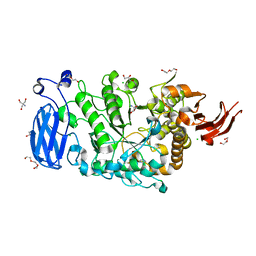 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltopentaose | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | 登録日 | 2019-02-12 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.981 Å) | | 主引用文献 | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1XD4
 
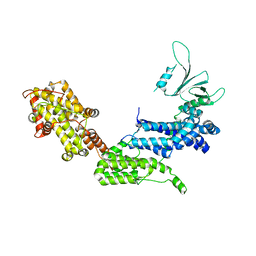 | | Crystal structure of the DH-PH-cat module of Son of Sevenless (SOS) | | 分子名称: | Son of sevenless protein homolog 1 | | 著者 | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | 登録日 | 2004-09-03 | | 公開日 | 2004-11-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
1XD2
 
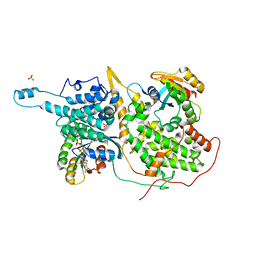 | | Crystal Structure of a ternary Ras:SOS:Ras*GDP complex | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | 登録日 | 2004-09-03 | | 公開日 | 2004-11-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
6JFJ
 
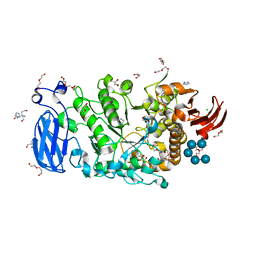 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltohexaose and alpha-cyclodextrin | | 分子名称: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | 登録日 | 2019-02-09 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.932 Å) | | 主引用文献 | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7MOQ
 
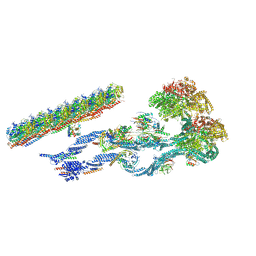 | | The structure of the Tetrahymena thermophila outer dynein arm on doublet microtubule | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Docking complex 1 protein, ... | | 著者 | Kubo, S, Yang, S.K, Ichikawa, M, Bui, K.H. | | 登録日 | 2021-05-03 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Remodeling and activation mechanisms of outer arm dyneins revealed by cryo-EM.
Embo Rep., 22, 2021
|
|
1XDV
 
 | | Experimentally Phased Structure of Human the Son of Sevenless protein at 4.1 Ang. | | 分子名称: | Son of sevenless protein homolog 1 | | 著者 | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | 登録日 | 2004-09-08 | | 公開日 | 2004-11-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
6JHF
 
 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | 登録日 | 2019-02-18 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
