1PXR
 
 | | Structure of Pro50Ala mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-06 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
1PY6
 
 | | Bacteriorhodopsin crystallized from bicells | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-08 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
1Q5I
 
 | | Crystal structure of bacteriorhodopsin mutant P186A crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Yohannan, S, Faham, S, Yang, D, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-08-07 | | Release date: | 2004-01-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q5J
 
 | | Crystal structure of bacteriorhodopsin mutant P91A crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Yohannan, S, Faham, S, Yang, D, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-08-07 | | Release date: | 2004-01-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6O2L
 
 | |
5DNK
 
 | | The structure of PKMT1 from Rickettsia prowazekii in complex with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
5DOO
 
 | | The structure of PKMT2 from Rickettsia typhi | | Descriptor: | CALCIUM ION, protein lysine methyltransferase 2 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-11 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.133 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
3WXZ
 
 | | The structure of the I375F mutant of CsyB | | Descriptor: | Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
3WY0
 
 | | The I375W mutant of CsyB complexed with CoA-SH | | Descriptor: | COENZYME A, Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
3WXY
 
 | | Crystal structure of CsyB complexed with CoA-SH | | Descriptor: | COENZYME A, Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
8ILS
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 17 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4BH5
 
 | | LytM domain of EnvC, an activator of cell wall amidases in Escherichia coli | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Morlot, C, Peters, N.T, Yang, D.C, Uehara, T, Vernet, T, Bernhardt, T.G. | | Deposit date: | 2013-03-29 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure-Function Analysis of the Lytm Domain of Envc, an Activator of Cell Wall Remodeling at the Escherichia Coli Division Site.
Mol.Microbiol., 89, 2013
|
|
5DPL
 
 | | The structure of PKMT2 from Rickettsia typhi in complex with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein lysine methyltransferase 2 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
5DO0
 
 | | The structure of PKMT1 from Rickettsia prowazekii | | Descriptor: | protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
1TN0
 
 | | Structure of bacterorhodopsin mutant A51P | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Yohannan, S, Yang, D, Faham, S, Boulting, G, Whitelegge, J, Bowie, J.U. | | Deposit date: | 2004-06-11 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Proline substitutions are not easily accommodated in a membrane protein
J.Mol.Biol., 341, 2004
|
|
1TN5
 
 | | Structure of bacterorhodopsin mutant K41P | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Yohannan, S, Yang, D, Faham, S, Boulting, G, Whitelegge, J, Bowie, J.U. | | Deposit date: | 2004-06-11 | | Release date: | 2004-10-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proline substitutions are not easily accommodated in a membrane protein
J.Mol.Biol., 341, 2004
|
|
3HAN
 
 | |
1EZO
 
 | | GLOBAL FOLD OF MALTODEXTRIN BINDING PROTEIN COMPLEXED WITH BETA-CYCLODEXTRIN | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Mueller, G.A, Choy, W.Y, Yang, D, Forman-Kay, J.D, Venters, R.A, Kay, L.E. | | Deposit date: | 2000-05-11 | | Release date: | 2001-05-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein.
J.Mol.Biol., 300, 2000
|
|
2KAP
 
 | | Solution structure of DLC1-SAM | | Descriptor: | Rho GTPase-activating protein 7 | | Authors: | Yang, S, Yang, D. | | Deposit date: | 2008-11-12 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Characterization of DLC1-SAM equilibrium unfolding at the amino acid residue level
Biochemistry, 48, 2009
|
|
8JIU
 
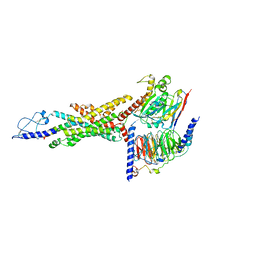 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1XAV
 
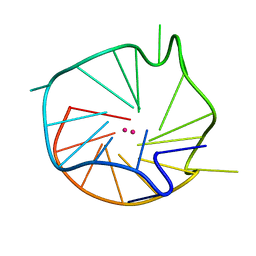 | | Major G-quadruplex structure formed in human c-MYC promoter, a monomeric parallel-stranded quadruplex | | Descriptor: | 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3', POTASSIUM ION | | Authors: | Ambrus, A, Chen, D, Dai, J, Jones, R.A, Yang, D. | | Deposit date: | 2004-08-26 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the biologically relevant G-Quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization.
Biochemistry, 44, 2005
|
|
1X95
 
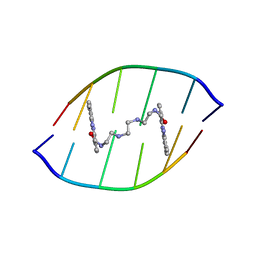 | | Solution structure of the DNA-hexamer ATGCAT complexed with DNA Bis-intercalating Anticancer Drug XR5944 (MLN944) | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, 5'-D(*AP*TP*GP*CP*AP*T)-3' | | Authors: | Dai, J, Punchihewa, C, Mistry, P, Ooi, A.T, Yang, D. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Novel DNA bis-intercalation by MLN944, a potent clinical bisphenazine anticancer drug.
J.Biol.Chem., 279, 2004
|
|
5H3N
 
 | |
5H3M
 
 | |
6NEB
 
 | | MYC Promoter G-Quadruplex with 1:6:1 loop length | | Descriptor: | DNA (27-MER) | | Authors: | Dickerhoff, J, Onel, B, Chen, L, Chen, Y, Yang, D. | | Deposit date: | 2018-12-17 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a MYC Promoter G-Quadruplex with 1:6:1 Loop Length.
Acs Omega, 4, 2019
|
|
