4MG2
 
 | | ALKBH2 F102A cross-linked to undamaged dsDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA-1, DNA-2, ... | | Authors: | Chen, B, Gan, J, Yang, C.G. | | Deposit date: | 2013-08-28 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The complex structures of ALKBH2 mutants cross-linked to
dsDNA reveal the conformational swing of β-hairpin
Sci China Chem, 57, 2014
|
|
5XVS
 
 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), LITHIUM ION, ... | | Authors: | Ko, T.P, Hsieh, T.J, Chen, S.C, Wu, S.C, Guan, H.H, Yang, C.H, Chen, C.J, Chen, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
4NES
 
 | | Crystal structure of Methanocaldococcus jannaschii UDP-GlcNAc 2-epimerase in complex with UDP-GlcNAc and UDP | | Descriptor: | UDP-N-acetylglucosamine 2-epimerase, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Chen, S.C, Yang, C.S, Huang, C.H, Chen, Y. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures of the archaeal UDP-GlcNAc 2-epimerase from Methanocaldococcus jannaschii reveal a conformational change induced by UDP-GlcNAc.
Proteins, 82, 2014
|
|
7ESG
 
 | |
5ZLR
 
 | | Structure of NeuC | | Descriptor: | GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), LITHIUM ION, SULFATE ION | | Authors: | Ko, T.P, Hsieh, T.J, Yang, C.S, Chen, Y. | | Deposit date: | 2018-03-29 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
6KKP
 
 | |
8Y54
 
 | |
5ZLT
 
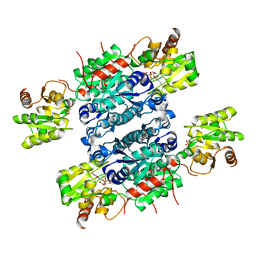 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | Descriptor: | GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), SULFATE ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Ko, T.P, Hsieh, T.J, Yang, C.S, Chen, Y. | | Deposit date: | 2018-03-29 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
8HY8
 
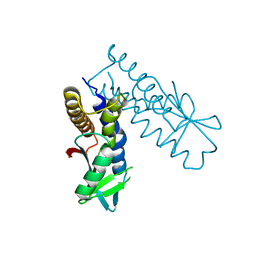 | | Bacterial STING from Epilithonimonas lactis | | Descriptor: | CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
2NOV
 
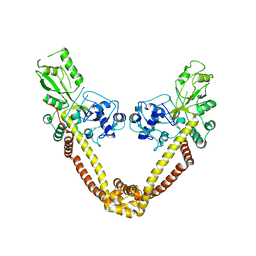 | | Breakage-reunion domain of S.pneumoniae topo IV: crystal structure of a gram-positive quinolone target | | Descriptor: | DNA topoisomerase 4 subunit A | | Authors: | Laponogov, I, Veselkov, D.A, Sohi, M.K, Pan, X.S, Achari, A, Yang, C, Ferrara, J.D, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Breakage-Reunion Domain of Streptococcus pneumoniae Topoisomerase IV: Crystal Structure of a Gram-Positive Quinolone Target.
PLoS ONE, 2, 2007
|
|
8XHW
 
 | | Haloquadratum walsbyi middle rhodopsin mutant - D84N | | Descriptor: | Bacteriorhodopsin-II-like protein, RETINAL | | Authors: | Ko, L.N, Lim, G.Z, Li, G.Y, Chen, J.C, Yang, C.S. | | Deposit date: | 2023-12-18 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of Haloquadratum walsbyi middle rhodopsin mutant D84N at 2.9 Angstroms resolution.
To Be Published
|
|
7VP9
 
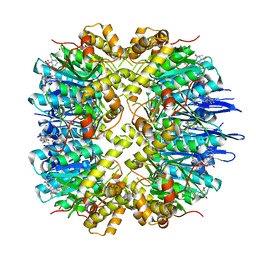 | | Crystal structure of human ClpP in complex with ZG111 | | Descriptor: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Wang, P.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Aberrant human ClpP activation disturbs mitochondrial proteome homeostasis to suppress pancreatic ductal adenocarcinoma.
Cell Chem Biol, 29, 2022
|
|
8HYN
 
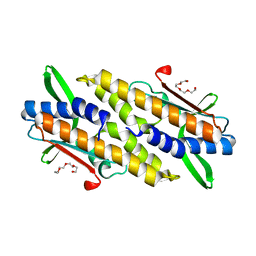 | | Bacterial STING from Riemerella anatipestifer | | Descriptor: | CD-NTase-associated protein 12, TETRAETHYLENE GLYCOL | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-07 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8JSJ
 
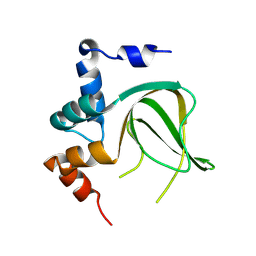 | |
2JE3
 
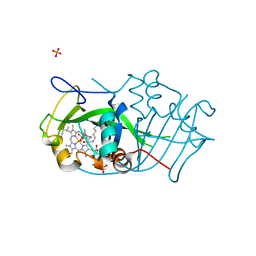 | | Cytochrome P460 from Nitrosomonas europaea - probable physiological form | | Descriptor: | CYTOCHROME P460, HEME C, PHOSPHATE ION | | Authors: | Pearson, A.R, Elmore, B.O, Yang, C, Ferrara, J.D, Hooper, A.B, Wilmot, C.M. | | Deposit date: | 2007-01-13 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Cytochrome P460 of Nitrosomonas Europaea Reveals a Novel Cytochrome Fold and Heme-Protein Cross-Link.
Biochemistry, 46, 2007
|
|
2JE2
 
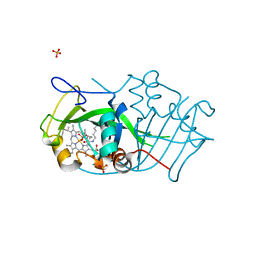 | | Cytochrome P460 from Nitrosomonas europaea - probable nonphysiological oxidized form | | Descriptor: | CYTOCHROME P460, HEME C, PHOSPHATE ION | | Authors: | Pearson, A.R, Elmore, B.O, Yang, C, Ferrara, J.D, Hooper, A.B, Wilmot, C.M. | | Deposit date: | 2007-01-13 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Cytochrome P460 of Nitrosomonas Europaea Reveals a Novel Cytochrome Fold and Heme-Protein Cross-Link.
Biochemistry, 46, 2007
|
|
8HWJ
 
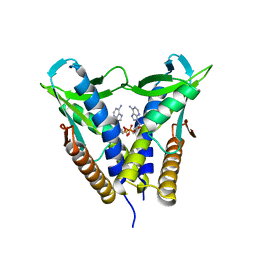 | | Bacterial STING from Epilithonimonas lactis in complex with 3'3'-c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HWI
 
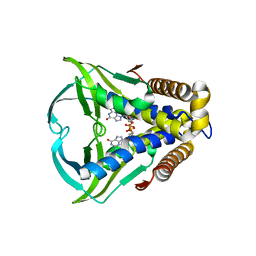 | | Bacterial STING from Larkinella arboricola in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HYK
 
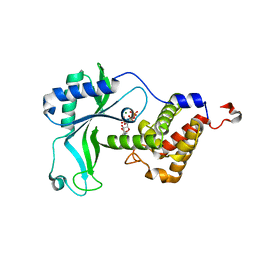 | | CD-NTase EfCdnE in complex with intermediate pppU[2'-5']p | | Descriptor: | EfCdnE, MAGNESIUM ION, [[(2R,3R,4R,5R)-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
8IML
 
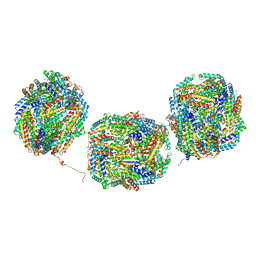 | | Rs2I-Rs2II, Rs1I-Rs1II, RbI-RbII cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster D) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMJ
 
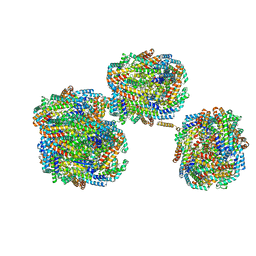 | | A'1-A'2, A'3-A'4, B1-B2, C1-C2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster B) | | Descriptor: | ApcA2, ApcB2, ApcB3, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMN
 
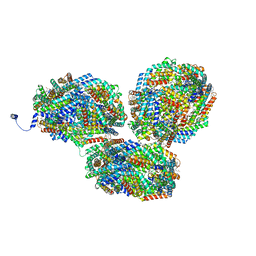 | | Rt1I-Rt1II, Rt2'I-Rt2'II, Rt3I-Rt3II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster F) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMM
 
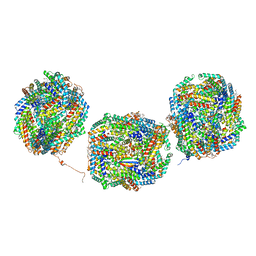 | | Rs2'I-Rs2'II, Rs1'I-Rs1'II, Rb'I-Rb'II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster E) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMO
 
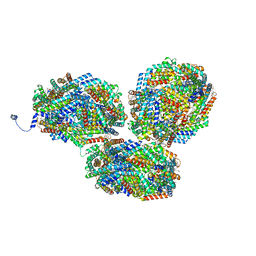 | | Rt1'I-Rt1'II, Rt2I-Rt2II, Rt3'I-Rt3'II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster G) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8HY9
 
 | | Bacterial STING from Riemerella anatipestifer in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.462 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
