6AAC
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with mAzZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AB1
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oAzZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-azidophenyl)methoxycarbonylamino]hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6ABL
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oBrZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-bromophenyl)methoxycarbonylamino]hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-22 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6ABM
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with pTmdZLys | | Descriptor: | (2S)-2-azanyl-6-[[4-[3-(trifluoromethyl)-1,2-diazirin-3-yl]phenyl]methoxycarbonylamino]hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-22 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.368 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAZ
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with pNO2ZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AB8
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with ZLys | | Descriptor: | (2S)-2-azanyl-6-(phenylmethoxycarbonylamino)hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-20 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AB2
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oClZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-chlorophenyl)methoxycarbonylamino]hexanoic acid, 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
3GH5
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3GH7
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3GH4
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 | | Descriptor: | ACETIC ACID, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
2CYB
 
 | | Crystal structure of Tyrosyl-tRNA Synthetase complexed with L-tyrosine from Archaeoglobus fulgidus | | Descriptor: | TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2006
|
|
2CYA
 
 | | Crystal structure of tyrosyl-tRNA synthetase from Aeropyrum pernix | | Descriptor: | SULFATE ION, Tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Murayama, K, Chen, L, Liu, Z.J, Wang, B.C, Kuroishi, C, Kuramitsu, S, Terada, T, Bessho, Y, Shirouzu, M, Sekine, S.I, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2005
|
|
2CYC
 
 | | Crystal structure of Tyrosyl-tRNA Synthetase complexed with L-tyrosine from Pyrococcus horikoshii | | Descriptor: | TYROSINE, tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Sakamoto, K, Terada, T, Shirouzu, M, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2006
|
|
5XHF
 
 | | Crystal structure of Trastuzumab Fab fragment bearing p-azido-L-phenylalanine | | Descriptor: | polypeptide (H chain), polypeptide (L chain) | | Authors: | Kuratani, M, Yanagisawa, T, Sakamoto, K, Yokoyama, S. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Extensive Survey of Antibody Invariant Positions for Efficient Chemical Conjugation Using Expanded Genetic Codes.
Bioconjug. Chem., 28, 2017
|
|
5XHG
 
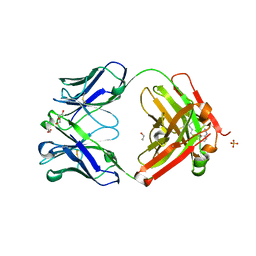 | | Crystal structure of Trastuzumab Fab fragment bearing Ne-(o-azidobenzyloxycarbonyl)-L-lysine | | Descriptor: | (2-azidophenyl)methyl hydrogen carbonate, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuratani, M, Yanagisawa, T, Sakamoto, K, Yokoyama, S. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Extensive Survey of Antibody Invariant Positions for Efficient Chemical Conjugation Using Expanded Genetic Codes.
Bioconjug. Chem., 28, 2017
|
|
5WXI
 
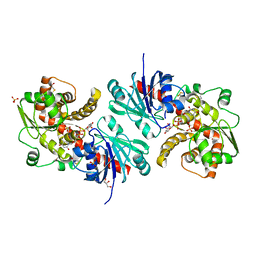 | | EarP bound with dTDP-rhamnose (soaked) | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, BETA-MERCAPTOETHANOL, EarP, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXJ
 
 | | Apo EarP | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, GLYCEROL, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXK
 
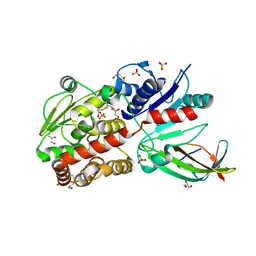 | | EarP bound with domain I of EF-P | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, Elongation factor P, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
8YFR
 
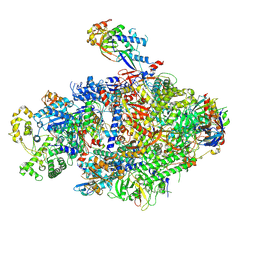 | | Cryo EM structure of Komagataella phaffii Rat1-Rai1 complex bound within the RNAPII cleft | | Descriptor: | 5'-3' exoribonuclease, DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Murayama, Y, Yanagisawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2024-02-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
8YFQ
 
 | | Cryo EM structure of Komagataella phaffii RNAPII-Rat1-Rai1 pre-termination complex | | Descriptor: | 5'-3' exoribonuclease, DNA (90-mer), DNA-directed RNA polymerase subunit, ... | | Authors: | Murayama, Y, Yanagisawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2024-02-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
5XVR
 
 | |
2ZUE
 
 | | Crystal structure of Pyrococcus horikoshii arginyl-tRNA synthetase complexed with tRNA(Arg) and an ATP analog (ANP) | | Descriptor: | Arginyl-tRNA synthetase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Konno, M, Sumida, T, Uchikawa, E, Mori, Y, Yanagisawa, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modeling of tRNA-assisted mechanism of Arg activation based on a structure of Arg-tRNA synthetase, tRNA, and an ATP analog (ANP)
Febs J., 276, 2009
|
|
2ZUF
 
 | | Crystal structure of Pyrococcus horikoshii arginyl-tRNA synthetase complexed with tRNA(Arg) | | Descriptor: | Arginyl-tRNA synthetase, tRNA-Arg | | Authors: | Konno, M, Sumida, T, Uchikawa, E, Mori, Y, Yanagisawa, T, Sekine, S, Yokoyama, S. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modeling of tRNA-assisted mechanism of Arg activation based on a structure of Arg-tRNA synthetase, tRNA, and an ATP analog (ANP)
Febs J., 276, 2009
|
|
3A5Z
 
 | | Crystal structure of Escherichia coli GenX in complex with elongation factor P | | Descriptor: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Elongation factor P, Putative lysyl-tRNA synthetase | | Authors: | Sumida, T, Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-08-17 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3A5Y
 
 | | Crystal structure of GenX from Escherichia coli in complex with lysyladenylate analog | | Descriptor: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Putative lysyl-tRNA synthetase | | Authors: | Sumida, T, Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-08-17 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P.
Nat.Struct.Mol.Biol., 17, 2010
|
|
