3TAT
 
 | | TYROSINE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, TYROSINE AMINOTRANSFERASE | | Authors: | Ko, T.P, Yang, W.Z, Wu, S.P, Tsai, H, Yuan, H.S. | | Deposit date: | 1998-08-12 | | Release date: | 1999-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of the Escherichia coli tyrosine aminotransferase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2AOQ
 
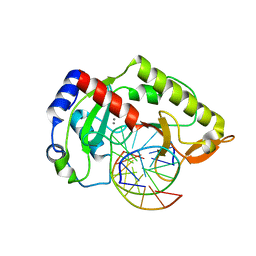 | | Crystal structure of MutH-unmethylated DNA complex | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*AP*TP*CP*AP*TP*GP*C)-3', CALCIUM ION, DNA mismatch repair protein mutH | | Authors: | Lee, J.Y, Chang, J, Joseph, N, Ghirlando, R, Rao, D.N, Yang, W. | | Deposit date: | 2005-08-13 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage.
Mol.Cell, 20, 2005
|
|
2AGO
 
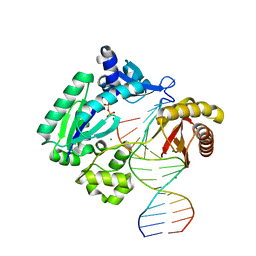 | | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*G)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Ling, H, Yang, W. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis.
Embo J., 24, 2005
|
|
2AGP
 
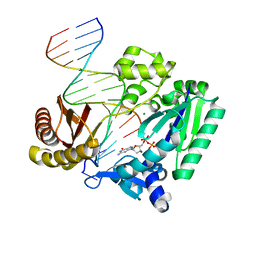 | | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*(DOC)-3'), ... | | Authors: | Ling, H, Yang, W. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis.
Embo J., 24, 2005
|
|
1ZBI
 
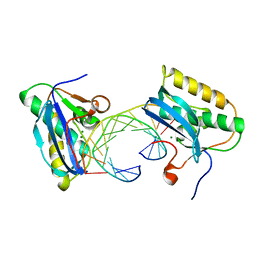 | | Bacillus halodurans RNase H catalytic domain mutant D132N in complex with 12-mer RNA/DNA hybrid | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3', 5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3', MAGNESIUM ION, ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis.
Cell(Cambridge,Mass.), 121, 2005
|
|
1ZBF
 
 | | Crystal structure of B. halodurans RNase H catalytic domain mutant D132N | | Descriptor: | SULFATE ION, ribonuclease H-related protein | | Authors: | Nowotny, M, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis.
Cell(Cambridge,Mass.), 121, 2005
|
|
3RJY
 
 | | Crystal Structure of Hyperthermophilic Endo-beta-1,4-glucanase in complex with substrate | | Descriptor: | Endoglucanase FnCel5A, PHOSPHATE ION, alpha-D-glucopyranose | | Authors: | Zheng, B, Yang, W, Zhao, X, Wang, Y, Lou, Z, Rao, Z, Feng, Y. | | Deposit date: | 2011-04-15 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of hyperthermophilic Endo-beta-1,4-glucanase: Implications for catalytic mechanism and thermostability.
J.Biol.Chem., 287, 2012
|
|
2A2K
 
 | | Crystal Structure of an active site mutant, C473S, of Cdc25B Phosphatase Catalytic Domain | | Descriptor: | CHLORIDE ION, M-phase inducer phosphatase 2, SULFATE ION | | Authors: | Sohn, J, Parks, J, Buhrman, G, Brown, P, Kristjansdottir, K, Safi, A, Yang, W, Edelsbrunner, H, Rudolph, J. | | Deposit date: | 2005-06-22 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Experimental Validation of the Docking Orientation of Cdc25 with Its Cdk2-CycA Protein Substrate.
Biochemistry, 44, 2005
|
|
3RJX
 
 | | Crystal Structure of Hyperthermophilic Endo-Beta-1,4-glucanase | | Descriptor: | Endoglucanase FnCel5A | | Authors: | Zheng, B.S, Yang, W, Zhao, X.Y, Wang, Y.G, Lou, Z.Y, Rao, Z.H, Feng, Y. | | Deposit date: | 2011-04-15 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of hyperthermophilic endo-beta-1,4-glucanase: implications for catalytic mechanism and thermostability.
J.Biol.Chem., 287, 2012
|
|
3RFE
 
 | | Crystal structure of glycoprotein GPIb ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Platelet glycoprotein Ib beta chain, ... | | Authors: | McEwan, P.A, Yang, W, Carr, K.H, Mo, X, Zheng, X, Li, R, Emsley, J. | | Deposit date: | 2011-04-06 | | Release date: | 2011-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.245 Å) | | Cite: | Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the structures of GPIbbeta and a GPIbbeta/GPIX chimer
Blood, 118, 2011
|
|
5D45
 
 | | Crystal Structure of FABP4 in complex with 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5YWG
 
 | | Crystal structure of Arabidopsis thaliana HPPD complexed with Mesotrione | | Descriptor: | 2-[(4-methylsulfonyl-2-nitro-phenyl)-oxidanyl-methylidene]cyclohexane-1,3-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, W.C. | | Deposit date: | 2017-11-29 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
1M7S
 
 | | Crystal Structure Analysis of Catalase CatF of Pseudomonas syringae | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Carpena, X, Soriano, M, Klotz, M.G, Duckworth, H.W, Donald, L.J, Melik-Adamyan, W, Fita, I, Loewen, P.C. | | Deposit date: | 2002-07-22 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Clade 1 catalase, CatF of Pseudomonas syringae, at 1.8 A resolution
Proteins, 50, 2003
|
|
4TLG
 
 | | Crystal structure of SEC14-like protein 4 (SEC14L4) | | Descriptor: | 1,2-ETHANEDIOL, SEC14-like protein 4, UNDECANOIC ACID | | Authors: | Vollmar, M, Kopec, J, Kiyani, W, Shrestha, L, Sorrell, F, Krojer, T, Williams, E, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of SEC14-like protein 4 (SEC14L4)
To Be Published
|
|
5D47
 
 | | Crystal Structure of FABP4 in complex with 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl] propanoic acid | | Descriptor: | 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl]propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D48
 
 | | Crystal Structure of FABP4 in complex with 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy) phenyl]-1H-indol-1-yl}propanoic acid | | Descriptor: | 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy)phenyl]-1H-indol-1-yl}propanoic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5XGK
 
 | | Crystal structure of Arabidopsis thaliana 4-hydroxyphenylpyruvate dioxygenase (AtHPPD) complexed with its substrate 4-hydroxyphenylpyruvate acid (HPPA) | | Descriptor: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, 4-hydroxyphenylpyruvate dioxygenase, ACETATE ION, ... | | Authors: | Yang, G.F, Yang, W.C, Lin, H.Y. | | Deposit date: | 2017-04-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of 4-Hydroxyphenylpyruvate Dioxygenase in Complex with Substrate Reveals a New Starting Point for Herbicide Discovery.
Res, 2019, 2019
|
|
1XDT
 
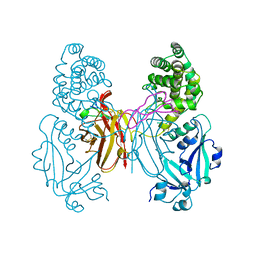 | | COMPLEX OF DIPHTHERIA TOXIN AND HEPARIN-BINDING EPIDERMAL GROWTH FACTOR | | Descriptor: | DIPHTHERIA TOXIN, HEPARIN-BINDING EPIDERMAL GROWTH FACTOR | | Authors: | Louie, G.V, Yang, W, Bowman, M.E, Choe, S. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex of diphtheria toxin with an extracellular fragment of its receptor.
Mol.Cell, 1, 1997
|
|
4B3O
 
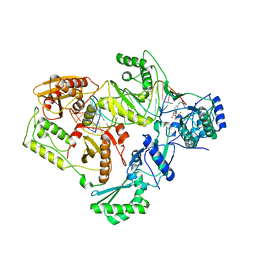 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, 5'-D(*CP*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP *TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3', 5'-R(*AP*UP*GP*AP*3DRP*GP*GP*CP*CP*AP*CP*AP*AP*UP*AP *AP*CP*UP*AP*UP*AP*GP*GP*CP*AP*UP*A)-3', ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4B3Q
 
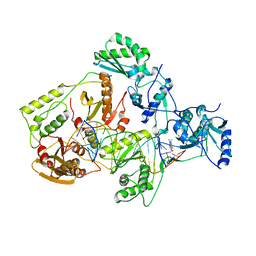 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, P51 RT, PRIMER DNA, ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
1GGF
 
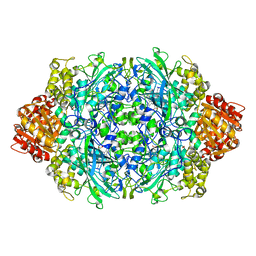 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, VARIANT HIS128ASN, COMPLEX WITH HYDROGEN PEROXIDE. | | Descriptor: | CATALASE HPII, HYDROGEN PEROXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-21 | | Release date: | 2000-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
4Y00
 
 | | Crystal Structure of Human TDP-43 RRM1 Domain with D169G Mutation in Complex with an Unmodified Single-stranded DNA | | Descriptor: | DNA (5'-D(P*TP*TP*GP*AP*GP*CP*GP*T)-3'), TAR DNA-binding protein 43 | | Authors: | Chiang, C.H, Kuo, P.H, Yang, W.Z, Yuan, H.S. | | Deposit date: | 2015-02-05 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Sci Rep, 6, 2016
|
|
2QKK
 
 | | Human RNase H catalytic domain mutant D210N in complex with 14-mer RNA/DNA hybrid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*G)-3', ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Ghirlando, R, Cerritelli, S.M, Crouch, R.J, Yang, W. | | Deposit date: | 2007-07-11 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Mol.Cell, 28, 2007
|
|
5AK8
 
 | | Structure of C351A mutant of Porphyromonas gingivalis peptidylarginine deiminase | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, ARGININE, ... | | Authors: | Kopec, J, Montgomery, A, Shrestha, L, Kiyani, W, Nowak, R, Burgess-Brown, N, Venables, P.J, Yue, W.W. | | Deposit date: | 2015-03-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Porphyromonas Gingivalis Peptidylarginine Deiminase: Implications for Autoimmunity in Rheumatoid Arthritis.
Ann.Rheum.Dis., 75, 2016
|
|
