8JRD
 
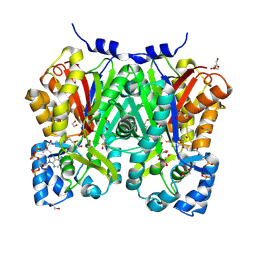 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waki, T, Imaizumi, R, Nakata, S, Yanai, T, Takeshita, K, Sakai, N, Kataoka, K, Yamamoto, M, Nakayama, T, Yamashita, S. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into catalytic promiscuity of chalcone synthase from Glycine max (L.) Merr.: Coenzyme A-induced alteration of product specificity.
Biochem.Biophys.Res.Commun., 718, 2024
|
|
6IHU
 
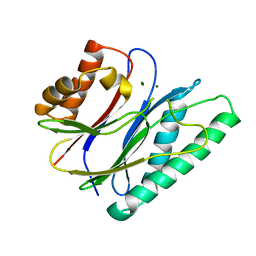 | |
6IHS
 
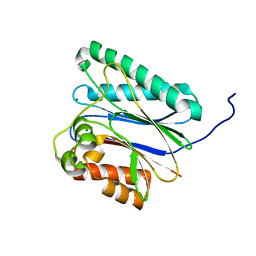 | |
6IHR
 
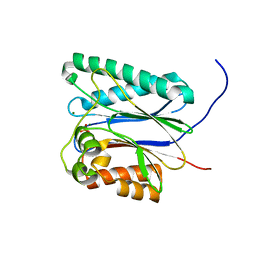 | |
6IHV
 
 | |
6IHT
 
 | |
6IHL
 
 | | Crystal structure of bacterial serine phosphatase | | Descriptor: | MAGNESIUM ION, Phosphorylated protein phosphatase | | Authors: | Yang, C.-G, yang, T. | | Deposit date: | 2018-09-30 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Structural Insight into the Mechanism of Staphylococcus aureus Stp1 Phosphatase.
Acs Infect Dis., 5, 2019
|
|
6IHW
 
 | |
3MFP
 
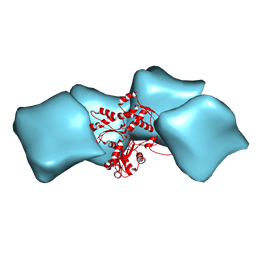 | | Atomic model of F-actin based on a 6.6 angstrom resolution cryoEM map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Fujii, T, Iwane, A.H, Yanagida, T, Namba, K. | | Deposit date: | 2010-04-03 | | Release date: | 2010-09-29 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Direct visualization of secondary structures of F-actin by electron cryomicroscopy
Nature, 467, 2010
|
|
5B42
 
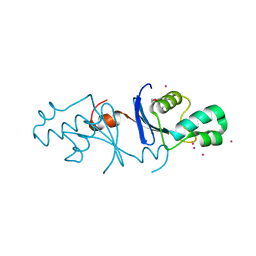 | | Crystal structure of the C-terminal endonuclease domain of Aquifex aeolicus MutL. | | Descriptor: | CADMIUM ION, DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL | | Authors: | Fukui, K, Baba, S, Kumasaka, T, Yano, T. | | Deposit date: | 2016-03-30 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Features and Functional Dependency on beta-Clamp Define Distinct Subfamilies of Bacterial Mismatch Repair Endonuclease MutL
J.Biol.Chem., 291, 2016
|
|
3GH4
 
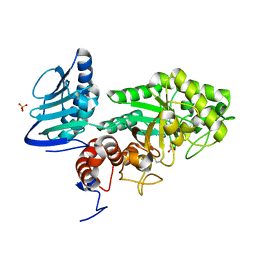 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 | | Descriptor: | ACETIC ACID, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3GH5
 
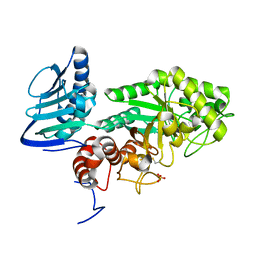 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3GH7
 
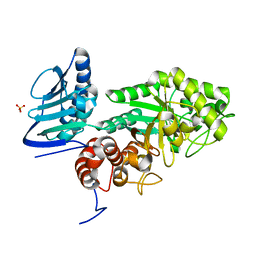 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
1CZC
 
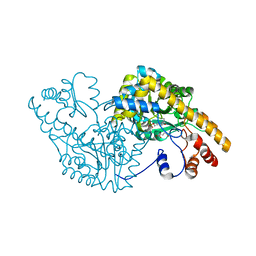 | | ASPARTATE AMINOTRANSFERASE MUTANT ATB17/139S/142N WITH GLUTARIC ACID | | Descriptor: | GLUTARIC ACID, PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Oue, S, Yano, T, Kagamiyama, H. | | Deposit date: | 1999-09-02 | | Release date: | 2000-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cocrystallization of a mutant aspartate aminotransferase with a C5-dicarboxylic substrate analog: structural comparison with the enzyme-C4-dicarboxylic analog complex.
J.Biochem.(Tokyo), 127, 2000
|
|
7DFU
 
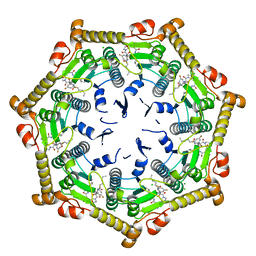 | |
7DFT
 
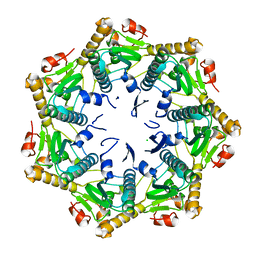 | | Crystal structure of Xanthomonas oryzae ClpP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CHLORIDE ION | | Authors: | Yang, C.-G, Yang, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dysregulation of ClpP by Small-Molecule Activators Used Against Xanthomonas oryzae pv. oryzae Infections.
J.Agric.Food Chem., 69, 2021
|
|
6INI
 
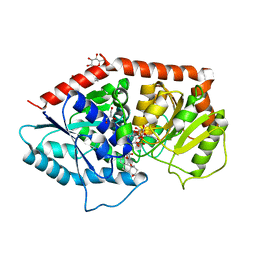 | | a glycosyltransferase complex with UDP and the product | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
6INF
 
 | | a glycosyltransferase complex with UDP | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
8H1G
 
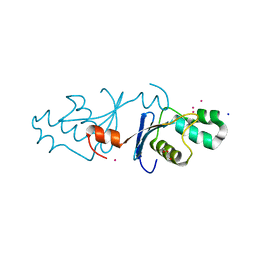 | |
8X36
 
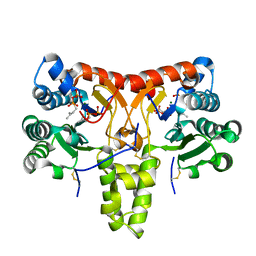 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form B) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8X35
 
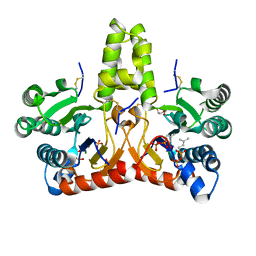 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8X37
 
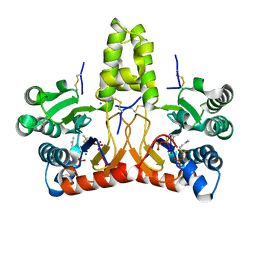 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
3HR4
 
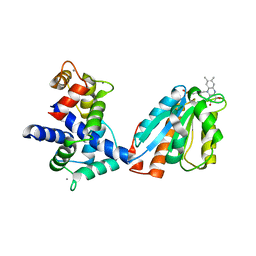 | | Human iNOS Reductase and Calmodulin Complex | | Descriptor: | CALCIUM ION, Calmodulin, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Xia, C, Misra, I, Iyanaki, T, Kim, J.J.K. | | Deposit date: | 2009-06-08 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of Interdomain Interactions by CaM in Inducible Nitric Oxide Synthase
J.Biol.Chem., 2009
|
|
8H1F
 
 | |
8H1E
 
 | |
