6AB1
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oAzZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-azidophenyl)methoxycarbonylamino]hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6ABL
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oBrZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-bromophenyl)methoxycarbonylamino]hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-22 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAN
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with mEtZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6ABM
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with pTmdZLys | | Descriptor: | (2S)-2-azanyl-6-[[4-[3-(trifluoromethyl)-1,2-diazirin-3-yl]phenyl]methoxycarbonylamino]hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-22 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.368 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAZ
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with pNO2ZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AB8
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with ZLys | | Descriptor: | (2S)-2-azanyl-6-(phenylmethoxycarbonylamino)hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-20 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAD
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with mTmdZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-[({3-[3-(trifluoromethyl)-3H-diaziren-3-yl]phenyl}methoxy)carbonyl]-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAQ
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with BCNLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-({[(1R,8S,9s)-bicyclo[6.1.0]non-4-yn-9-yl]methoxy}carbonyl)-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AB2
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oClZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-chlorophenyl)methoxycarbonylamino]hexanoic acid, 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
2EL9
 
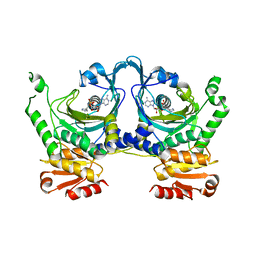 | | Crystal structure of E.coli Histidyl-tRNA synthetase complexed with a histidyl-adenylate analogue | | Descriptor: | 5'-O-[(L-HISTIDYLAMINO)SULFONYL]ADENOSINE, Histidyl-tRNA synthetase | | Authors: | Yanagisawa, T, Si, S.Y, Matsuno, M, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of E.coli Histidyl-tRNA synthetase complexed with a histidyl-adenylate analogue
To be published
|
|
7X8M
 
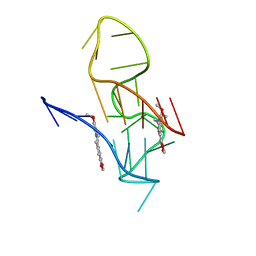 | | NMR Solution Structure of the 2:1 Berberine-KRAS-G4 Complex | | Descriptor: | BERBERINE, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8O
 
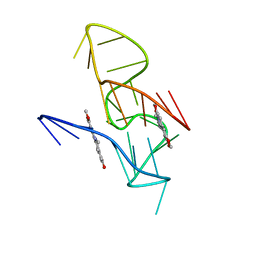 | | NMR Solution Structure of the 2:1 Coptisine-KRAS-G4 Complex | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8N
 
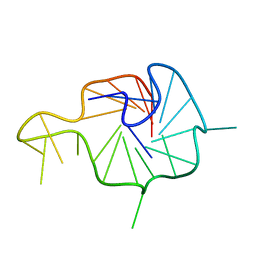 | | NMR Solution Structure of the Wild-type Bulge-containing KRAS-G4 | | Descriptor: | DNA (24-mer) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
8H4X
 
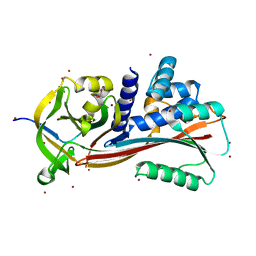 | | Structure of cleaved Serpin B9 | | Descriptor: | 1,2-ETHANEDIOL, Serpin B9, ZINC ION | | Authors: | Zhou, A, Yan, T. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Expression, purification and crystallization of cleaved Serpin B9
To Be Published
|
|
7X1Z
 
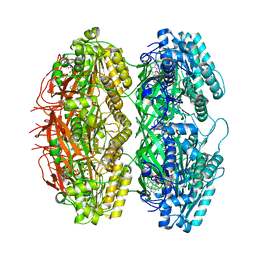 | | Structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
7X1Y
 
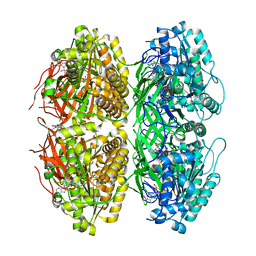 | | Structure of the phosphorylation-site double mutant S431A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
1ZVM
 
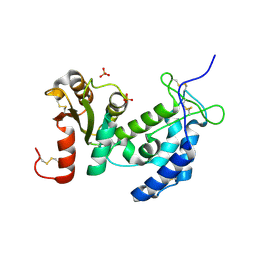 | | Crystal structure of human CD38: cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase | | Descriptor: | ADP-ribosyl cyclase 1, SULFATE ION | | Authors: | Shi, W, Yang, T, Almo, S.C, Schramm, V.L, Sauve, A. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human CD38: Cyclic-ADP-ribosyl synthetase/NAD+ glycohydrolase
To be Published
|
|
7YG7
 
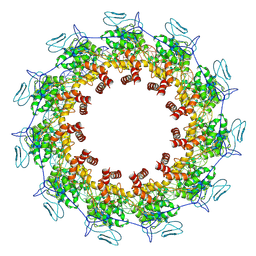 | | Structure of the Spring Viraemia of Carp Virus ribonucleoprotein Complex | | Descriptor: | Nucleoprotein, RNA (99-mer) | | Authors: | Liu, B, Wang, Z.X, Yang, T, Yu, D.Q, Ouyang, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Spring Viraemia of Carp Virus Ribonucleoprotein Complex Reveals Its Assembly Mechanism and Application in Antiviral Drug Screening.
J.Virol., 97, 2023
|
|
8KGA
 
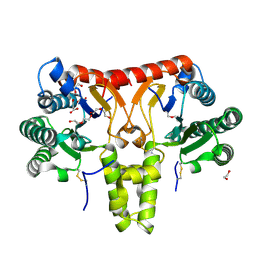 | | SlNDPS1-AtcPT4 Chimera | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dimethylallylcistransferase CPT1, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Biosynthesising various rubber-like polymers by reconstituting prenyltransferases on Hevea rubber particles: molecular and structural bases of the versatile system
To Be Published
|
|
8KGB
 
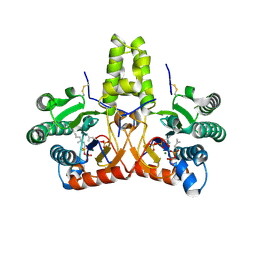 | | SlNDPS1-AtcPT4 Chimera complexed with GSPP, Mg2+, and IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Dimethylallylcistransferase CPT1, chloroplastic,Dehydrodolichyl diphosphate synthase 2, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A versatile system for enzymatic synthesis of natural and unnatural polyisoprenoids on rubber particles
To Be Published
|
|
5WXI
 
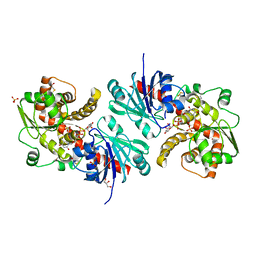 | | EarP bound with dTDP-rhamnose (soaked) | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, BETA-MERCAPTOETHANOL, EarP, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXJ
 
 | | Apo EarP | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, GLYCEROL, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
8IYT
 
 | | Crystal Structure of Serine Palmitoyltransferase complexed with D-methylserine | | Descriptor: | (2~{R})-2-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Racemization of the substrate and product by serine palmitoyltransferase from Sphingobacterium multivorum yields two enantiomers of the product from d-serine.
J.Biol.Chem., 300, 2024
|
|
8IYP
 
 | | Crystal structure of serine palmitoyltransferase soaked in 190 mM D-serine solution | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Racemization of the substrate and product by serine palmitoyltransferase from Sphingobacterium multivorum yields two enantiomers of the product from d-serine.
J.Biol.Chem., 300, 2024
|
|
8ECY
 
 | | cryoEM structure of bovine bestrophin-2 and glutamine synthetase complex | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Owji, A.P, Kittredge, A.K, Yang, T. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Bestrophin-2 and glutamine synthetase form a complex for glutamate release.
Nature, 611, 2022
|
|
