9CTT
 
 | | Best1 + GABA closed state | | Descriptor: | Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-07-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | GAD65 tunes the functions of Best1 as a GABA receptor and a neurotransmitter conducting channel.
Nat Commun, 15, 2024
|
|
9CTR
 
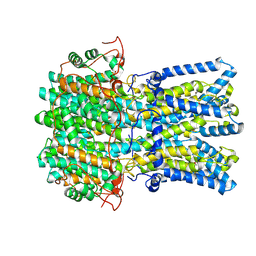 | | Best1 + GABA intermediate state 1 | | Descriptor: | Bestrophin-1, CALCIUM ION, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-07-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | GAD65 tunes the functions of Best1 as a GABA receptor and a neurotransmitter conducting channel.
Nat Commun, 15, 2024
|
|
7X1Z
 
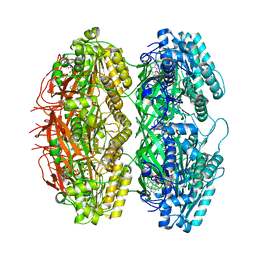 | | Structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
7X1Y
 
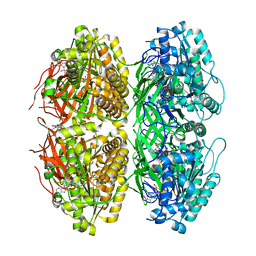 | | Structure of the phosphorylation-site double mutant S431A/T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock oscillator protein KaiC, MAGNESIUM ION | | Authors: | Han, X, Zhang, D.L, Hong, L, Yu, D.Q, Wu, Z.L, Yang, T, Rust, M.J, Tu, Y.H, Ouyang, Q. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Determining subunit-subunit interaction from statistics of cryo-EM images: observation of nearest-neighbor coupling in a circadian clock protein complex
Nat Commun, 14, 2023
|
|
7YG7
 
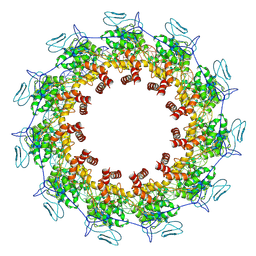 | | Structure of the Spring Viraemia of Carp Virus ribonucleoprotein Complex | | Descriptor: | Nucleoprotein, RNA (99-mer) | | Authors: | Liu, B, Wang, Z.X, Yang, T, Yu, D.Q, Ouyang, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Spring Viraemia of Carp Virus Ribonucleoprotein Complex Reveals Its Assembly Mechanism and Application in Antiviral Drug Screening.
J.Virol., 97, 2023
|
|
1CZC
 
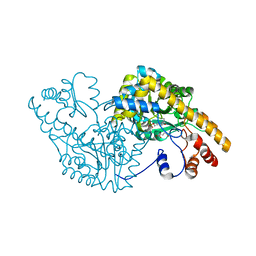 | | ASPARTATE AMINOTRANSFERASE MUTANT ATB17/139S/142N WITH GLUTARIC ACID | | Descriptor: | GLUTARIC ACID, PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Oue, S, Yano, T, Kagamiyama, H. | | Deposit date: | 1999-09-02 | | Release date: | 2000-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cocrystallization of a mutant aspartate aminotransferase with a C5-dicarboxylic substrate analog: structural comparison with the enzyme-C4-dicarboxylic analog complex.
J.Biochem.(Tokyo), 127, 2000
|
|
1CZE
 
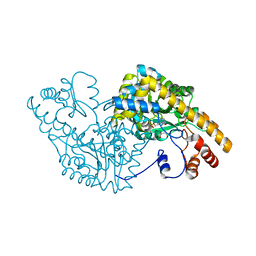 | | ASPARTATE AMINOTRANSFERASE MUTANT ATB17/139S/142N WITH SUCCINIC ACID | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SUCCINIC ACID | | Authors: | Okamoto, A, Oue, S, Yano, T, Kagamiyama, H. | | Deposit date: | 1999-09-02 | | Release date: | 2000-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cocrystallization of a mutant aspartate aminotransferase with a C5-dicarboxylic substrate analog: structural comparison with the enzyme-C4-dicarboxylic analog complex.
J.Biochem.(Tokyo), 127, 2000
|
|
8ECY
 
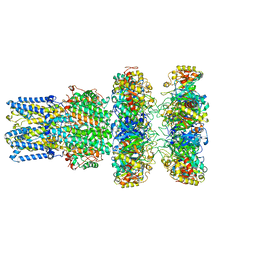 | | cryoEM structure of bovine bestrophin-2 and glutamine synthetase complex | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Owji, A.P, Kittredge, A.K, Yang, T. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Bestrophin-2 and glutamine synthetase form a complex for glutamate release.
Nature, 611, 2022
|
|
6IHT
 
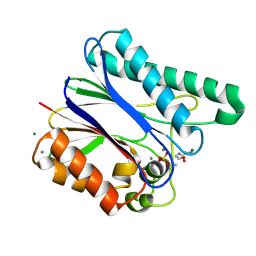 | |
6IHL
 
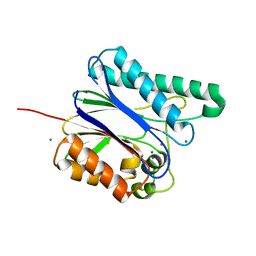 | | Crystal structure of bacterial serine phosphatase | | Descriptor: | MAGNESIUM ION, Phosphorylated protein phosphatase | | Authors: | Yang, C.-G, yang, T. | | Deposit date: | 2018-09-30 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Structural Insight into the Mechanism of Staphylococcus aureus Stp1 Phosphatase.
Acs Infect Dis., 5, 2019
|
|
6IHW
 
 | |
6IHU
 
 | |
6IHV
 
 | |
6IHR
 
 | |
6IHS
 
 | |
6VX7
 
 | | bestrophin-2 Ca2+-bound state (5 mM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX8
 
 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX9
 
 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX6
 
 | | bestrophin-2 Ca2+-bound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX5
 
 | | bestrophin-2 Ca2+- unbound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2VPY
 
 | | Polysulfide reductase with bound quinone inhibitor, pentachlorophenol (PCP) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VPW
 
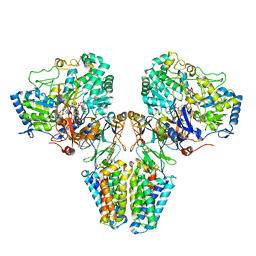 | | Polysulfide reductase with bound menaquinone | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VPZ
 
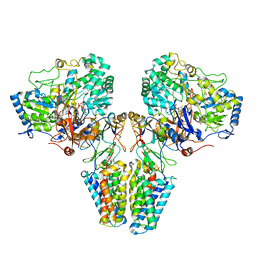 | | Polysulfide reductase native structure | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VPX
 
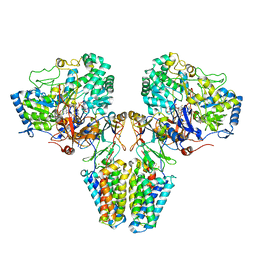 | | Polysulfide reductase with bound quinone (UQ1) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5OCV
 
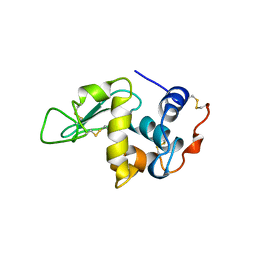 | | A Rare Lysozyme Crystal Form Solved Using High-Redundancy 3D Electron Diffraction Data from Micron-Sized Needle Shaped Crystals | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Xu, H, Lebrette, H, Yang, T, Srinivas, V, Hovmoller, S, Hogbom, M, Zou, X. | | Deposit date: | 2017-07-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | Cite: | A Rare Lysozyme Crystal Form Solved Using Highly Redundant Multiple Electron Diffraction Datasets from Micron-Sized Crystals.
Structure, 26, 2018
|
|
