3D2B
 
 | |
3D2A
 
 | |
1Y8I
 
 | | Horse methemoglobin low salt, PH 7.0 (98% relative humidity) | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
1Y8K
 
 | | Horse methemoglobin low salt, PH 7.0 (88% relative humidity) | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
1Y8H
 
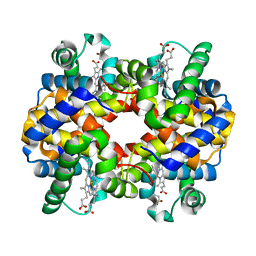 | | HORSE METHEMOGLOBIN LOW SALT, PH 7.0 | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
1QF6
 
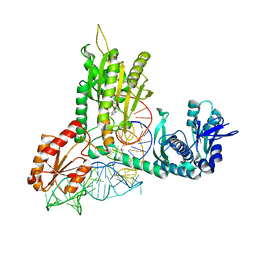 | | STRUCTURE OF E. COLI THREONYL-TRNA SYNTHETASE COMPLEXED WITH ITS COGNATE TRNA | | Descriptor: | ADENOSINE MONOPHOSPHATE, THREONINE TRNA, THREONYL-TRNA SYNTHETASE, ... | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 1999-04-06 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site
Cell(Cambridge,Mass.), 97, 1999
|
|
6Y7C
 
 | | Early cytoplasmic yeast pre-40S particle (purified with Tsr1 as bait) | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Plassart, L, Plisson-Chastang, C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Good Vibrations: Structural Remodeling of Maturing Yeast Pre-40S Ribosomal Particles Followed by Cryo-Electron Microscopy.
Molecules, 25, 2020
|
|
8AU1
 
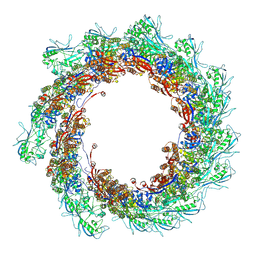 | | Jumbo Phage phi-kp24 tail outer sheath | | Descriptor: | Putative tail sheath protein | | Authors: | Ouyang, R, Briegel, A. | | Deposit date: | 2022-08-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
3AAY
 
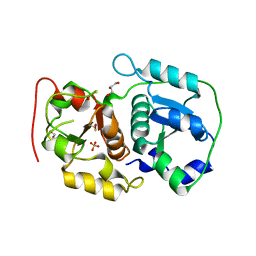 | | Crystal structure of probable thiosulfate sulfurtransferase CYSA3 (RV3117) from Mycobacterium tuberculosis: orthorhombic form | | Descriptor: | GLYCEROL, Putative thiosulfate sulfurtransferase, SULFATE ION | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
3AAX
 
 | | Crystal structure of probable thiosulfate sulfurtransferase cysa3 (RV3117) from Mycobacterium tuberculosis: monoclinic FORM | | Descriptor: | Putative thiosulfate sulfurtransferase | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
7UMI
 
 | | Importin a1 bound to Cp183-CTD | | Descriptor: | HBV-NLS, Importin subunit alpha-1 | | Authors: | Yang, R, Cingolani, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for nuclear import of hepatitis B virus (HBV) nucleocapsid core.
Sci Adv, 10, 2024
|
|
2I6U
 
 | | Crystal Structure of Ornithine Carbamoyltransferase complexed with Carbamoyl Phosphate and L-Norvaline from Mycobacterium tuberculosis (Rv1656) at 2.2 A | | Descriptor: | NORVALINE, Ornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Sankaranarayanan, R, Moradian, F, Cherney, L.T, Garen, C, Cherney, M.M, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-08-29 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of ornithine carbamoyltransferase from Mycobacterium tuberculosis and its ternary complex with carbamoyl phosphate and L-norvaline reveal the enzyme's catalytic mechanism
J.Mol.Biol., 375, 2008
|
|
7UCJ
 
 | | Mammalian 80S translation initiation complex with mRNA and Harringtonine | | Descriptor: | 18S rRNA, 28s rRNA, 40S ribosomal protein S10, ... | | Authors: | Yang, R, Arango, D, Sturgill, D, Oberdoerffer, S. | | Deposit date: | 2022-03-16 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine.
Mol.Cell, 82, 2022
|
|
7UCK
 
 | | 80S translation initiation complex with ac4c(-1) mRNA and Harringtonine | | Descriptor: | 18S rRNA, 28s rRNA, 40S ribosomal protein S10, ... | | Authors: | Yang, R, Arango, D, Sturgill, D, Oberdoerffer, S. | | Deposit date: | 2022-03-16 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine.
Mol.Cell, 82, 2022
|
|
2P2G
 
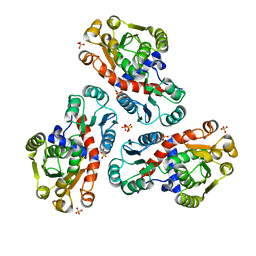 | | Crystal Structure of Ornithine Carbamoyltransferase from Mycobacterium Tuberculosis (Rv1656): Orthorhombic Form | | Descriptor: | Ornithine carbamoyltransferase, SULFATE ION | | Authors: | Sankaranarayanan, R, Cherney, M.M, Cherney, L.T, Garen, C, Moradian, F, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structures of ornithine carbamoyltransferase from Mycobacterium tuberculosis and its ternary complex with carbamoyl phosphate and L-norvaline reveal the enzyme's catalytic mechanism.
J.Mol.Biol., 375, 2008
|
|
1EVL
 
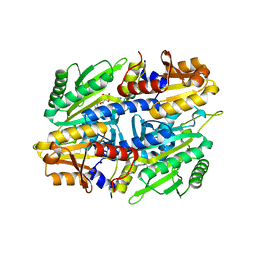 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE WITH A THREONYL ADENYLATE ANALOG | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 2000-04-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase.
Nat.Struct.Biol., 7, 2000
|
|
1FYF
 
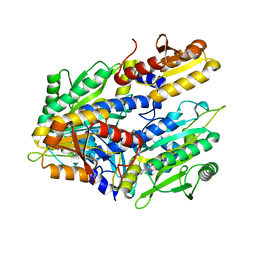 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE COMPLEXED WITH A SERYL ADENYLATE ANALOG | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2000-09-29 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem.
Cell(Cambridge,Mass.), 103, 2000
|
|
1EVK
 
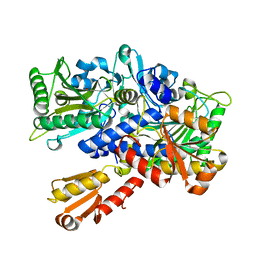 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE WITH THE LIGAND THREONINE | | Descriptor: | THREONINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 2000-04-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase.
Nat.Struct.Biol., 7, 2000
|
|
8W6C
 
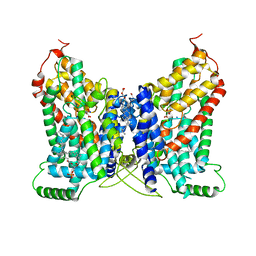 | | CryoEM structure of NaDC1 with Citrate | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CHOLESTEROL HEMISUCCINATE, CITRIC ACID, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Dai, L, Zhang, Y, Shen, Y, Chen, Y, Shi, T, Yang, H, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8EQJ
 
 | | Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like membrane environment, MSP1D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8EQU
 
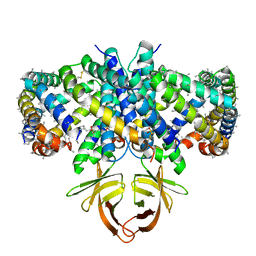 | | Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like environment, Saposin A nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein, Saposin A, ... | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8EQT
 
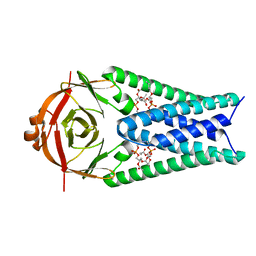 | | Structure of SARS-CoV-2 Orf3a in plasma membrane-like environment, MSP1D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8EQS
 
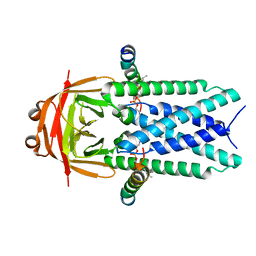 | | Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like environment, MSP1D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apolipoprotein A-I, ORF3a protein | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
6NPS
 
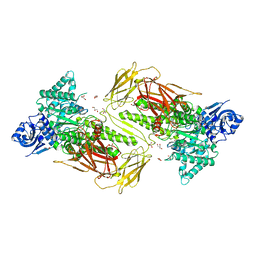 | | Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus | | Descriptor: | AxyAgu115A, CHLORIDE ION, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Yan, R, Master, E, Savchenko, A. | | Deposit date: | 2019-01-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural characterization of the family GH115 alpha-glucuronidase from Amphibacillus xylanus yields insight into its coordinated action with alpha-arabinofuranosidases.
N Biotechnol, 2021
|
|
7RW6
 
 | | BORF2-APOBEC3Bctd Complex | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3B, Maltose/maltodextrin-binding periplasmic protein,Ribonucleoside-diphosphate reductase large subunit, ZINC ION | | Authors: | Shaban, N.M, Yan, R, Shi, K, McLellan, J.S, Yu, Z, Harris, R.S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure of the EBV ribonucleotide reductase BORF2 and mechanism of APOBEC3B inhibition.
Sci Adv, 8, 2022
|
|
