6MR1
 
 | | RbcS-like subdomain of CcmM | | 分子名称: | CHLORIDE ION, COBALT (II) ION, Carbon dioxide concentrating mechanism protein, ... | | 著者 | Ryan, P, Kimber, M.S. | | 登録日 | 2018-10-11 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | The small RbcS-like domains of the beta-carboxysome structural protein CcmM bind RubisCO at a site distinct from that binding the RbcS subunit.
J. Biol. Chem., 294, 2019
|
|
5GIK
 
 | | Modulation of the affinity of a HIV-1 capsid-directed ankyrin towards its viral target through critical amino acid editing | | 分子名称: | Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant -S45Y, GLYCEROL | | 著者 | Chuankhayan, P, Saoin, S, Wisitponchai, T, Intachai, K, Chupradit, K, Moonmuang, S, Kitidee, K, Nangola, S, Sanghiran, L.V, Hong, S.S, Boulanger, P, Tayapiwatana, C, Chen, C.J. | | 登録日 | 2016-06-24 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Modulation of the affinity of a HIV-1 capsid-directed ankyrintowards its viral target through critical amino acid editing
To Be Published
|
|
7EXF
 
 | | Crystal structure of wild-type from Arabidopsis thaliana complexed with Galactose | | 分子名称: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-galactopyranose | | 著者 | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | 登録日 | 2021-05-27 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXH
 
 | | Crystal structure of D383A mutant from Arabidopsis thaliana complexed with Galactinol. | | 分子名称: | Probable galactinol--sucrose galactosyltransferase 6, galactinol | | 著者 | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | 登録日 | 2021-05-27 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXJ
 
 | | Crystal structure of alkaline alpha-galctosidase D383A mutant from Arabidopsis thaliana complexed with Raffinose | | 分子名称: | Probable galactinol--sucrose galactosyltransferase 6, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | 著者 | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | 登録日 | 2021-05-27 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXR
 
 | | Crystal structure of alkaline alpha-galactosidase D383A mutant from Arabidopsis thaliana complexed with Stachyose. | | 分子名称: | Probable galactinol--sucrose galactosyltransferase 6, alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | 著者 | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | 登録日 | 2021-05-28 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXG
 
 | | Crystal structure of D383A mutant from Arabidopsis thaliana complexed with Galactose. | | 分子名称: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-galactopyranose | | 著者 | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | 登録日 | 2021-05-27 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXQ
 
 | | Crystal structure of alkaline alpha-galactosidase D383A mutant from Arabidopsis thaliana complexed with product-galactose and sucrose. | | 分子名称: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-D-galactopyranose | | 著者 | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | 登録日 | 2021-05-28 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4L7Q
 
 | | Crystal structure of gamma glutamyl hydrolase (wild-type) from zebrafish | | 分子名称: | GLYCEROL, Gamma-glutamyl hydrolase | | 著者 | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | 登録日 | 2013-06-14 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
4L8W
 
 | | Crystal structure of gamma glutamyl hydrolase (H218N) from zebrafish complex with MTX polyglutamate | | 分子名称: | D-GLUTAMIC ACID, Gamma-glutamyl hydrolase, METHOTREXATE | | 著者 | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | 登録日 | 2013-06-18 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
4L8Y
 
 | |
4L8F
 
 | | Crystal structure of gamma-glutamyl hydrolase (C108A) complex with MTX | | 分子名称: | Gamma-glutamyl hydrolase, METHOTREXATE | | 著者 | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | 登録日 | 2013-06-17 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
4L95
 
 | | Crystal structure of gamma glutamyl hydrolase (H218N) from zebrafish | | 分子名称: | GLYCEROL, Gamma-glutamyl hydrolase | | 著者 | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | 登録日 | 2013-06-18 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
7F65
 
 | | Bacetrial Cocaine Esterase with mutations T172R/G173Q/V116K/S117A/A51L, bound to benzoic acid | | 分子名称: | BENZOIC ACID, Cocaine esterase, SULFATE ION | | 著者 | Ouyang, P.F, Zhang, Y, Tong, J. | | 登録日 | 2021-06-24 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Computational Design and Crystal Structure of a Highly Efficient Benzoylecgonine Hydrolase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7WL3
 
 | | CVB5 expended empty particle | | 分子名称: | Capsid protein, Genome polyprotein | | 著者 | Yang, P, Wang, K. | | 登録日 | 2022-01-12 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Atomic Structures of Coxsackievirus B5 Provide Key Information on Viral Evolution and Survival.
J.Virol., 96, 2022
|
|
7XB2
 
 | |
7MN2
 
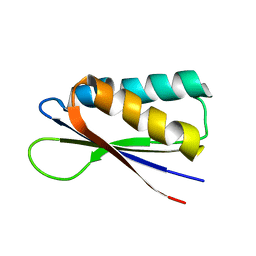 | | Rules for designing protein fold switches and their implications for the folding code | | 分子名称: | Sb2 | | 著者 | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | 登録日 | 2021-04-30 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MP7
 
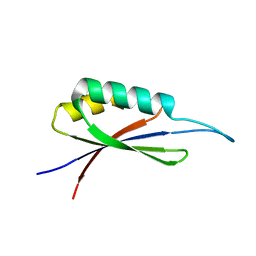 | | Rules for designing protein fold switches and their implications for the folding code | | 分子名称: | Sb3 | | 著者 | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | 登録日 | 2021-05-04 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MN1
 
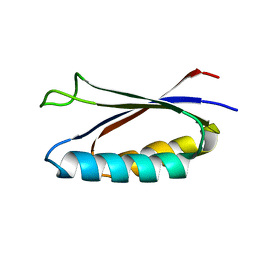 | | Rules for designing protein fold switches and their implications for the folding code | | 分子名称: | Sa1 | | 著者 | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | 登録日 | 2021-04-30 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MQ4
 
 | | Rules for designing protein fold switches and their implications for the folding code | | 分子名称: | Sb1 | | 著者 | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | 登録日 | 2021-05-05 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
1VKT
 
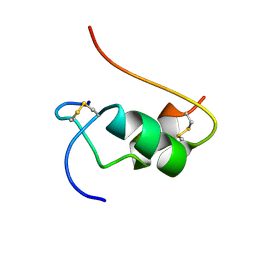 | | HUMAN INSULIN TWO DISULFIDE MODEL, NMR, 10 STRUCTURES | | 分子名称: | INSULIN | | 著者 | Hua, Q.X, Hu, S.Q, Frank, B.H, Jia, W.H, Chu, Y.C, Wang, S.H, Burke, G.T, Katsoyannis, P.G, Weiss, M.A. | | 登録日 | 1996-10-14 | | 公開日 | 1997-04-01 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mapping the functional surface of insulin by design: structure and function of a novel A-chain analogue.
J.Mol.Biol., 264, 1996
|
|
2FS1
 
 | | solution structure of PSD-1 | | 分子名称: | PSD-1 | | 著者 | He, Y, Rozak, D.A, Sari, N, Chen, Y, Bryan, P, Orban, J. | | 登録日 | 2006-01-20 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, dynamics, and stability variation in bacterial albumin binding modules: implications for species specificity.
Biochemistry, 45, 2006
|
|
5BUN
 
 | | Crystal structure of an antigenic outer membrane protein ST50 from Salmonella Typhi | | 分子名称: | Outer membrane protein, octyl beta-D-glucopyranoside | | 著者 | Yoshimura, M, Chuankhayan, P, Lin, C.C, Chen, N.C, Yang, M.C, Fun, H.K. | | 登録日 | 2015-06-04 | | 公開日 | 2015-12-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Crystal structure of an antigenic outer-membrane protein from Salmonella Typhi suggests a potential antigenic loop and an efflux mechanism.
Sci Rep, 5, 2015
|
|
8EXX
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and foscarnet (pre-translocation state) | | 分子名称: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | 著者 | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | 登録日 | 2022-10-26 | | 公開日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Viral DNA polymerase structures reveal mechanisms for drug selectivity
Cell(Cambridge,Mass.), 2024
|
|
8V1T
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and acyclovir triphosphate in closed conformation | | 分子名称: | ACYCLOVIR TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | 著者 | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | 登録日 | 2023-11-21 | | 公開日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Viral DNA polymerase structures reveal mechanisms for drug selectivity
Cell(Cambridge,Mass.), 2024
|
|
