1UG6
 
 | | Structure of beta-glucosidase at atomic resolution from thermus thermophilus HB8 | | Descriptor: | GLYCEROL, beta-glycosidase | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-12 | | Release date: | 2003-06-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structure of Beta-Glucosidase at Atomic Resolution from Thermus Thermophilus Hb8
To be Published
|
|
1UB7
 
 | |
1UB0
 
 | | Crystal Structure Analysis of Phosphomethylpyrimidine Kinase (ThiD) from Thermus Thermophilus Hb8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Phosphomethylpyrimidine Kinase, ... | | Authors: | Bagautdinov, B, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-26 | | Release date: | 2003-04-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure Analysis of Phosphomethylpyrimidine Kinase (ThiD) from Thermus Thermophilus Hb8
To be Published
|
|
1UFY
 
 | |
1UI9
 
 | | Crystal analysis of chorismate mutase from thermus thermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, chorismate mutase | | Authors: | Inagaki, E, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-15 | | Release date: | 2003-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of chorismate mutase from thermus thermophilus
To be Published
|
|
1UKW
 
 | | Crystal structure of medium-chain acyl-CoA dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | COBALT (II) ION, FLAVIN-ADENINE DINUCLEOTIDE, acyl-CoA dehydrogenase | | Authors: | Hamada, K, Ago, H, Kuramitsu, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-02 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus thermophilus medium-chain acyl-CoA dehydrogenase
To be published
|
|
1VEL
 
 | | Mycobacterium smegmatis Dps tetragonal form | | Descriptor: | CADMIUM ION, SODIUM ION, SULFATE ION, ... | | Authors: | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2004-04-01 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules
J.Mol.Biol., 339, 2004
|
|
2LY1
 
 | |
1VEQ
 
 | | Mycobacterium smegmatis Dps Hexagonal form | | Descriptor: | FE (III) ION, starvation-induced DNA protecting protein | | Authors: | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2004-04-03 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules
J.Mol.Biol., 339, 2004
|
|
1VEI
 
 | | Mycobacterium smegmatis Dps | | Descriptor: | FE (III) ION, SULFATE ION, starvation-induced DNA protecting protein | | Authors: | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray Analysis of Mycobacterium smegmatis Dps and a Comparative Study Involving Other Dps and Dps-like Molecules
J.Mol.Biol., 339, 2004
|
|
1V8E
 
 | |
2LH9
 
 | |
2LY2
 
 | |
1J3L
 
 | | Structure of the RNA-processing inhibitor RraA from Thermus thermophilis | | Descriptor: | CHLORIDE ION, Demethylmenaquinone Methyltransferase, MAGNESIUM ION | | Authors: | Rehse, P.H, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-04 | | Release date: | 2004-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the RNA-processing inhibitor RraA from Thermus thermophilis.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2O02
 
 | | Phosphorylation independent interactions between 14-3-3 and Exoenzyme S: from structure to pathogenesis | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, ExoS (416-430) peptide | | Authors: | Ottmann, C, Yasmin, L, Weyand, M, Hauser, A.R, Wittinghofer, A, Hallberg, B. | | Deposit date: | 2006-11-27 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphorylation-independent interaction between 14-3-3 and exoenzyme S: from structure to pathogenesis
Embo J., 26, 2007
|
|
2CVZ
 
 | |
2DCH
 
 | | Crystal structure of archaeal intron-encoded homing endonuclease I-Tsp061I | | Descriptor: | CHLORIDE ION, SULFATE ION, putative homing endonuclease | | Authors: | Nakayama, H, Tsuge, H, Shimamura, T, Miyano, M, Nomura, N, Sako, Y. | | Deposit date: | 2006-01-06 | | Release date: | 2006-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of a hyperthermophilic archaeal homing endonuclease, I-Tsp061I: contribution of cross-domain polar networks to thermostability.
J.Mol.Biol., 365, 2007
|
|
2CVD
 
 | | Crystal structure analysis of human hematopoietic prostaglandin D synthase complexed with HQL-79 | | Descriptor: | 4-(BENZHYDRYLOXY)-1-[3-(1H-TETRAAZOL-5-YL)PROPYL]PIPERIDINE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Aritake, K, Kado, Y, Inoue, T, Miyano, M, Urade, Y. | | Deposit date: | 2005-06-02 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of HQL-79, an Orally Selective Inhibitor of Human Hematopoietic Prostaglandin D Synthase.
J.Biol.Chem., 281, 2006
|
|
2CLI
 
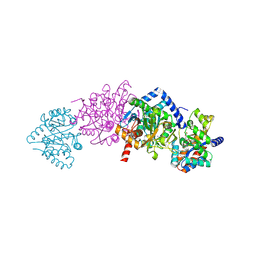 | | Tryptophan Synthase in complex with N-(4'- trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and Characterization of Allosteric Probes of Substrate Channeling in the Tryptophan Synthase Bienzyme Complex.
Biochemistry, 46, 2007
|
|
2CLH
 
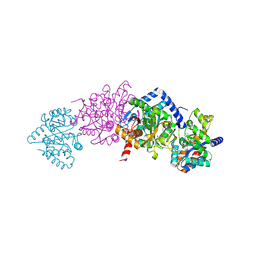 | | Tryptophan Synthase in complex with (naphthalene-2'-sulfonyl)-2-amino- 1-ethylphosphate (F19) | | Descriptor: | 2-[(2-NAPHTHYLSULFONYL)AMINO]ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and characterization of allosteric probes of substrate channeling in the tryptophan synthase bienzyme complex.
Biochemistry, 46, 2007
|
|
2DM6
 
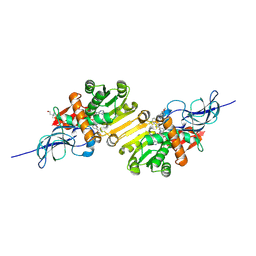 | | Crystal structure of anti-configuration of indomethacin and leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complex | | Descriptor: | INDOMETHACIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent leukotriene B4 12-hydroxydehydrogenase, ... | | Authors: | Hori, T, Ishijima, J, Yokomizo, T, Ago, H, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-20 | | Release date: | 2006-11-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of anti-configuration of indomethacin and leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complex reveals the structural basis of broad spectrum indomethacin efficacy
J.Biochem.(Tokyo), 140, 2006
|
|
1UIY
 
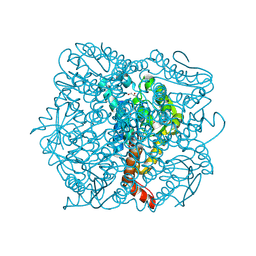 | | Crystal Structure of Enoyl-CoA Hydratase from Thermus Thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Enoyl-CoA Hydratase, GLYCEROL | | Authors: | Bagautdinov, B, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-24 | | Release date: | 2003-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
1UKK
 
 | | Structure of Osmotically Inducible Protein C from Thermus thermophilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Osmotically Inducible Protein C | | Authors: | Rehse, P.H, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-24 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Structure and Biochemical Analysis of the Thermus thermophilus Osmotically Inducible Protein C
J.MOL.BIOL., 338, 2004
|
|
2LVM
 
 | |
2K2W
 
 | | Second BRCT domain of NBS1 | | Descriptor: | Recombination and DNA repair protein | | Authors: | Xu, C, Cui, G, Botuyan, M, Mer, G. | | Deposit date: | 2008-04-14 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a second BRCT domain identified in the nijmegen breakage syndrome protein Nbs1 and its function in an MDC1-dependent localization of Nbs1 to DNA damage sites.
J.Mol.Biol., 381, 2008
|
|
