3RBB
 
 | | HIV-1 NEF protein in complex with engineered HCK SH3 domain | | Descriptor: | 1,2-ETHANEDIOL, Protein Nef, Tyrosine-protein kinase HCK | | Authors: | Horenkamp, F.A, Schulte, A, Weyand, M, Geyer, M. | | Deposit date: | 2011-03-29 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conformation of the Dileucine-Based Sorting Motif in HIV-1 Nef Revealed by Intermolecular Domain Assembly.
Traffic, 12, 2011
|
|
3MIU
 
 | | Structure of Banana Lectin-pentamannose complex | | Descriptor: | HEXANE-1,6-DIOL, Lectin, ZINC ION, ... | | Authors: | Sharma, A, Vijayan, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Influence of glycosidic linkage on the nature of carbohydrate binding in beta-prism I fold lectins: an X-ray and molecular dynamics investigation on banana lectin-carbohydrate complexes
Glycobiology, 21, 2011
|
|
5AHW
 
 | | Crystal structure of universal stress protein MSMEG_3811 in complex with cAMP | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Adolph, R.S, Kleinboelting, S, Weyand, M, Steegborn, C. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Universal Stress Protein (Usp) in Mycobacteria Binds Camp
J.Biol.Chem., 290, 2015
|
|
5VF0
 
 | |
5VWE
 
 | |
3MIT
 
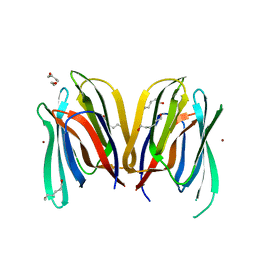 | | Structure of Banana lectin-alpha-D-mannose complex | | Descriptor: | HEXANE-1,6-DIOL, Lectin, ZINC ION, ... | | Authors: | Sharma, A, Vijayan, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Influence of glycosidic linkage on the nature of carbohydrate binding in beta-prism I fold lectins: an X-ray and molecular dynamics investigation on banana lectin-carbohydrate complexes
Glycobiology, 21, 2011
|
|
5BOH
 
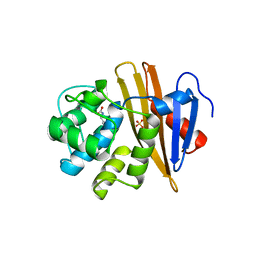 | |
1APA
 
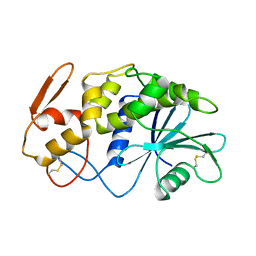 | | X-RAY STRUCTURE OF A POKEWEED ANTIVIRAL PROTEIN, CODED BY A NEW GENOMIC CLONE, AT 0.23 NM RESOLUTION. A MODEL STRUCTURE PROVIDES A SUITABLE ELECTROSTATIC FIELD FOR SUBSTRATE BINDING. | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Ago, H, Kataoka, J, Tsuge, H, Habuka, N, Inagaki, E, Noma, M, Miyano, M. | | Deposit date: | 1993-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of a pokeweed antiviral protein, coded by a new genomic clone, at 0.23 nm resolution. A model structure provides a suitable electrostatic field for substrate binding.
Eur.J.Biochem., 225, 1994
|
|
1CIW
 
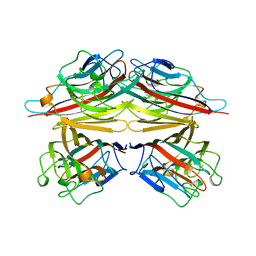 | | PEANUT LECTIN COMPLEXED WITH N-ACETYLLACTOSAMINE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-04-06 | | Release date: | 1999-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the complexes of peanut lectin with methyl-beta-galactose and N-acetyllactosamine and a comparative study of carbohydrate binding in Gal/GalNAc-specific legume lectins.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3SD4
 
 | |
2Q39
 
 | |
2Q2P
 
 | |
7V21
 
 | |
7V1Y
 
 | |
7V1Z
 
 | | human Serine beta-lactamase-like protein LACTB | | Descriptor: | Serine beta-lactamase-like protein LACTB, mitochondrial | | Authors: | Zhang, M.H, Yang, M.J. | | Deposit date: | 2021-08-07 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis for the catalytic activity of filamentous human serine beta-lactamase-like protein LACTB.
Structure, 30, 2022
|
|
7E3O
 
 | |
1J1Y
 
 | | Crystal Structure of PaaI from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PaaI protein | | Authors: | Kunishima, N, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-24 | | Release date: | 2004-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
3ED8
 
 | |
3LW1
 
 | | Binary complex of 14-3-3 sigma and p53 pT387-peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Schumacher, B, Mondry, J, Thiel, P, Weyand, M, Ottmann, C. | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure of the p53 C-terminus bound to 14-3-3: Implications for stabilization of the p53 tetramer
Febs Lett., 584, 2010
|
|
7WGW
 
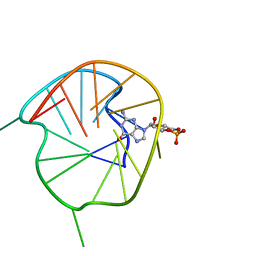 | | NMR Solution Structure of a cGMP Fill-in Vacancy G-quadruplex Formed in the Oxidized BLM Gene Promoter | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, DNA (20-MER) | | Authors: | Wang, K.B, Liu, Y, Li, Y, Li, J, Dickerhoff, J, Yang, M.H, Yang, D, Kong, L.Y. | | Deposit date: | 2021-12-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oxidative Damage Induces a Vacancy G-Quadruplex That Binds Guanine Metabolites: Solution Structure of a cGMP Fill-in Vacancy G-Quadruplex in the Oxidized BLM Gene Promoter.
J.Am.Chem.Soc., 144, 2022
|
|
7D14
 
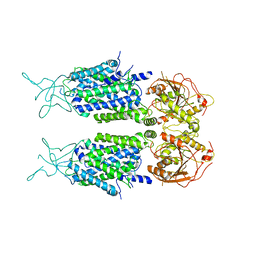 | | Mouse KCC2 | | Descriptor: | Solute carrier family 12 member 5 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-13 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
7D10
 
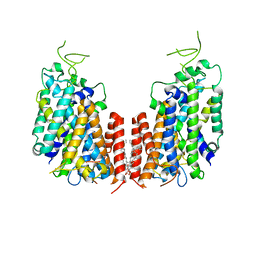 | | Human NKCC1 | | Descriptor: | PALMITIC ACID, Solute carrier family 12 member 2 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-12 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
3MIV
 
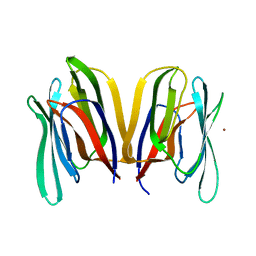 | | Structure of Banana lectin - Glc-alpha(1,2)-Glc complex | | Descriptor: | Lectin, ZINC ION | | Authors: | Sharma, A, Vijayan, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Influence of glycosidic linkage on the nature of carbohydrate binding in beta-prism I fold lectins: an X-ray and molecular dynamics investigation on banana lectin-carbohydrate complexes
Glycobiology, 21, 2011
|
|
8GAG
 
 | | Cannabinoid receptor 1-Gi complex with novel ligand | | Descriptor: | Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tummino, T.A, Iliopoulos-Tsoutsouvas, C, Braz, J.M, O'Brien, E.S, Krishna Kumar, K, Makriyannis, M, Basbaum, A.I, Shoichet, B.K. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cannabinoid receptor 1-Gi complex with novel ligand
To Be Published
|
|
1LQM
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
