3T5W
 
 | | 2ME modified human SOD1 | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Ihara, K, Yamaguchi, Y, Torigoe, H, Wakatsuki, S, Taniguchi, N, Suzuki, K, Fujiwara, N. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural switching of Cu,Zn-superoxide dismutases at loop VI: insights from the crystal structure of 2-mercaptoethanol-modified enzyme
Biosci.Rep., 32, 2012
|
|
5B1W
 
 | |
5B1X
 
 | |
5AZX
 
 | | Crystal structure of p24delta1 GOLD domain (native 1) | | Descriptor: | SULFATE ION, Transmembrane emp24 domain-containing protein 10 | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-09-14 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | 3D Structure and Interaction of p24 beta and p24 delta Golgi Dynamics Domains: Implication for p24 Complex Formation and Cargo Transport
J.Mol.Biol., 428, 2016
|
|
5AZW
 
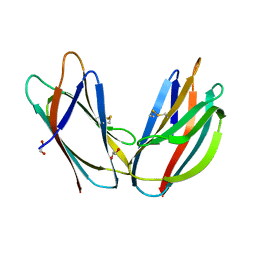 | | Crystal structure of p24beta1 GOLD domain | | Descriptor: | 1,2-ETHANEDIOL, Transmembrane emp24 domain-containing protein 2 | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 3D Structure and Interaction of p24 beta and p24 delta Golgi Dynamics Domains: Implication for p24 Complex Formation and Cargo Transport
J.Mol.Biol., 428, 2016
|
|
5AZY
 
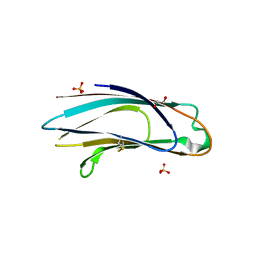 | | Crystal structure of p24delta1 GOLD domain (Native 2) | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Transmembrane emp24 domain-containing protein 10 | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 3D Structure and Interaction of p24 beta and p24 delta Golgi Dynamics Domains: Implication for p24 Complex Formation and Cargo Transport
J.Mol.Biol., 428, 2016
|
|
3WCR
 
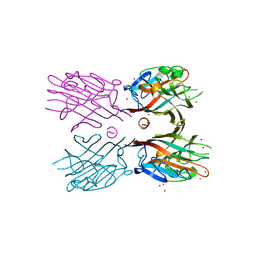 | | Crystal structure of plant lectin (ligand-free form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, Erythroagglutinin | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2013-05-31 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
3W1W
 
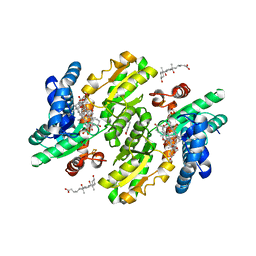 | | Protein-drug complex | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXYBENZOIC ACID, CHOLIC ACID, ... | | Authors: | Ishii, R, Gupta, V, Yamaguchi, Y, Handa, H, Nureki, O. | | Deposit date: | 2012-11-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Salicylic Acid induces mitochondrial injury by inhibiting ferrochelatase heme biosynthesis activity
Mol.Pharmacol., 84, 2013
|
|
3VQZ
 
 | | Crystal structure of metallo-beta-lactamase, SMB-1, in a complex with mercaptoacetic acid | | Descriptor: | Metallo-beta-lactamase, SODIUM ION, SULFANYLACETIC ACID, ... | | Authors: | Wachino, J, Yamaguchi, Y, Mori, S, Arakawa, Y, Shibayama, K. | | Deposit date: | 2012-04-02 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Subclass B3 Metallo-beta-Lactamase SMB-1 and the Mode of Inhibition by the Common Metallo- -Lactamase Inhibitor Mercaptoacetate
Antimicrob.Agents Chemother., 57, 2013
|
|
3VPE
 
 | | Crystal Structure of Metallo-beta-Lactamase SMB-1 | | Descriptor: | ACETATE ION, GLYCEROL, Metallo-beta-lactamase, ... | | Authors: | Wachino, J, Yamaguchi, Y, Mori, S, Arakawa, Y, Shibayama, K. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Subclass B3 Metallo-beta-Lactamase SMB-1 and the Mode of Inhibition by the Common Metallo- -Lactamase Inhibitor Mercaptoacetate
Antimicrob.Agents Chemother., 57, 2013
|
|
3WBQ
 
 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-2) | | Descriptor: | C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WOC
 
 | | Crystal structure of a prostate-specific WGA16 glycoprotein lectin, form II | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Garenaux, E, Kanagawa, M, Tsuchiyama, T, Hori, K, Kanazawa, T, Goshima, A, Chiba, M, Yasue, H, Ikeda, A, Yamaguchi, Y, Sato, C, Kitajima, K. | | Deposit date: | 2013-12-26 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery, Primary, and Crystal Structures and Capacitation-related Properties of a Prostate-derived Heparin-binding Protein WGA16 from Boar Sperm
J.Biol.Chem., 290, 2015
|
|
3WBP
 
 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-1) | | Descriptor: | 1,2-ETHANEDIOL, C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WWK
 
 | | Crystal structure of CLEC-2 in complex with rhodocytin | | Descriptor: | C-type lectin domain family 1 member B, Snaclec rhodocytin subunit alpha, Snaclec rhodocytin subunit beta | | Authors: | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | Deposit date: | 2014-06-20 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
3WBR
 
 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-3) | | Descriptor: | C-type lectin domain family 4 member C | | Authors: | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WSR
 
 | | Crystal structure of CLEC-2 in complex with O-glycosylated podoplanin | | Descriptor: | C-type lectin domain family 1 member B, Peptide from Podoplanin, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | Deposit date: | 2014-03-20 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
3VZE
 
 | |
3VY6
 
 | |
3VZF
 
 | |
3VY7
 
 | |
3VZG
 
 | |
5AV7
 
 | | Crystal structure of Calsepa lectin in complex with bisected glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl alpha-D-mannopyranoside, Lectin | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-06-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Atomic visualization of a flipped-back conformation of bisected glycans bound to specific lectins
Sci Rep, 6, 2016
|
|
1WXS
 
 | | Solution Structure of Ufm1, a ubiquitin-fold modifier | | Descriptor: | Ubiquitin-fold Modifier 1 | | Authors: | Sasakawa, H, Sakata, E, Yamaguchi, Y, Komatsu, M, Tatsumi, K, Kominami, E, Tanaka, K, Kato, K. | | Deposit date: | 2005-02-01 | | Release date: | 2006-04-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1
Biochem.Biophys.Res.Commun., 343, 2006
|
|
2D5U
 
 | |
2DPF
 
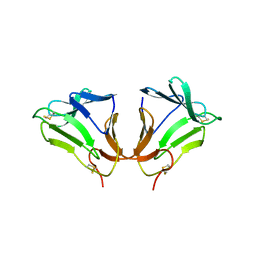 | | Crystal Structure of curculin1 homodimer | | Descriptor: | Curculin, SULFATE ION | | Authors: | Kurimoto, E, Suzuki, M, Amemiya, E, Yamaguchi, Y, Nirasawa, S, Shimba, N, Xu, N, Kashiwagi, T, Kawai, M, Suzuki, E, Kato, K. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Curculin Exhibits Sweet-tasting and Taste-modifying Activities through Its Distinct Molecular Surfaces.
J.Biol.Chem., 282, 2007
|
|
